Inherited SLP76 deficiency in humans causes severe combined immunodeficiency, neutrophil and platelet defects
- PMID: 33231617
- PMCID: PMC7690938
- DOI: 10.1084/jem.20201062
Inherited SLP76 deficiency in humans causes severe combined immunodeficiency, neutrophil and platelet defects
Abstract
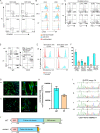
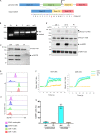
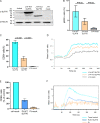
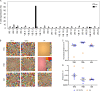
The T cell receptor (TCR) signaling pathway is an ensemble of numerous proteins that are crucial for an adequate immune response. Disruption of any protein involved in this pathway leads to severe immunodeficiency and unfavorable clinical outcomes. Here, we describe an infant with severe immunodeficiency who was found to have novel biallelic mutations in SLP76. SLP76 is a key protein involved in TCR signaling and in other hematopoietic pathways. Previous studies of this protein were performed using Jurkat-derived human leukemic T cell lines and SLP76-deficient mice. Our current study links this gene, for the first time, to a human immunodeficiency characterized by early-onset life-threatening infections, combined T and B cell immunodeficiency, severe neutrophil defects, and impaired platelet aggregation. Hereby, we characterized aspects of the patient's immune phenotype, modeled them with an SLP76-deficient Jurkat-derived T cell line, and rescued some consequences using ectopic expression of wild-type SLP76. Understanding human diseases due to SLP76 deficiency is helpful in explaining the mixed T cell and neutrophil defects, providing a guide for exploring human SLP76 biology.
© 2020 Lev et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures


References
-
- Bacchelli, C., Moretti F.A., Carmo M., Adams S., Stanescu H.C., Pearce K., Madkaikar M., Gilmour K.C., Nicholas A.K., Woods C.G., et al. . 2017. Mutations in linker for activation of T cells (LAT) lead to a novel form of severe combined immunodeficiency. J. Allergy Clin. Immunol. 139:634–642.e5. 10.1016/j.jaci.2016.05.036 - DOI - PubMed
-
- Bryceson, Y.T., Pende D., Maul-Pavicic A., Gilmour K.C., Ufheil H., Vraetz T., Chiang S.C., Marcenaro S., Meazza R., Bondzio I., et al. . 2012. A prospective evaluation of degranulation assays in the rapid diagnosis of familial hemophagocytic syndromes. Blood. 119:2754–2763. 10.1182/blood-2011-08-374199 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

