Chromosomal genome of Triplophysa bleekeri provides insights into its evolution and environmental adaptation
- PMID: 33231676
- PMCID: PMC7684707
- DOI: 10.1093/gigascience/giaa132
Chromosomal genome of Triplophysa bleekeri provides insights into its evolution and environmental adaptation
Abstract
Background: Intense stresses caused by high-altitude environments may result in noticeable genetic adaptions in native species. Studies of genetic adaptations to high elevations have been largely limited to terrestrial animals. How fish adapt to high-elevation environments is largely unknown. Triplophysa bleekeri, an endemic fish inhabiting high-altitude regions, is an excellent model to investigate the genetic mechanisms of adaptation to the local environment. Here, we assembled a chromosomal genome sequence of T. bleekeri, with a size of ∼628 Mb (contig and scaffold N50 of 3.1 and 22.9 Mb, respectively). We investigated the origin and environmental adaptation of T. bleekeri based on 21,198 protein-coding genes in the genome.
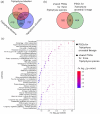
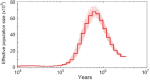
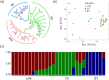
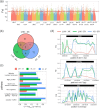
Results: Compared with fish species living at low altitudes, gene families associated with lipid metabolism and immune response were significantly expanded in the T. bleekeri genome. Genes involved in DNA repair exhibit positive selection for T. bleekeri, Triplophysa siluroides, and Triplophysa tibetana, indicating that adaptive convergence in Triplophysa species occurred at the positively selected genes. We also analyzed whole-genome variants among samples from 3 populations. The results showed that populations separated by geological and artificial barriers exhibited obvious differences in genetic structures, indicating that gene flow is restricted between populations.
Conclusions: These results will help us expand our understanding of environmental adaptation and genetic diversity of T. bleekeri and provide valuable genetic resources for future studies on the evolution and conservation of high-altitude fish species such as T. bleekeri.
Keywords: Triplophysa bleekeri; genetic adaptation; genome; population genomics.
© The Author(s) 2020. Published by Oxford University Press GigaScience.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Myers N, Mittermeier RA, Mittermeier CG, et al. Biodiversity hotspots for conservation priorities. Nature. 2000;403(6772):853. - PubMed
-
- Zhao Z, Li S. Extinction vs. rapid radiation: the juxtaposed evolutionary histories of coelotine spiders support the Eocene–Oligocene orogenesis of the Tibetan Plateau. Syst Biol. 2017;66(6):988–1006. - PubMed
-
- Beall CM. Adaptation to high altitude: phenotypes and genotypes. Annu Rev Anthropol. 2014;43:251–72.
-
- Monge C, Leonvelarde F. Physiological adaptation to high altitude: oxygen transport in mammals and birds. Physiol Rev. 1991;71(4):1135–72. - PubMed
-
- Wu T, Kayser B. High altitude adaptation in Tibetans. High Alt Med Biol. 2006;7(3):193–208. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

