GTRD: an integrated view of transcription regulation
- PMID: 33231677
- PMCID: PMC7778956
- DOI: 10.1093/nar/gkaa1057
GTRD: an integrated view of transcription regulation
Abstract
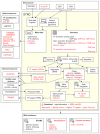
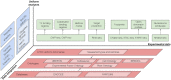
The Gene Transcription Regulation Database (GTRD; http://gtrd.biouml.org/) contains uniformly annotated and processed NGS data related to gene transcription regulation: ChIP-seq, ChIP-exo, DNase-seq, MNase-seq, ATAC-seq and RNA-seq. With the latest release, the database has reached a new level of data integration. All cell types (cell lines and tissues) presented in the GTRD were arranged into a dictionary and linked with different ontologies (BRENDA, Cell Ontology, Uberon, Cellosaurus and Experimental Factor Ontology) and with related experiments in specialized databases on transcription regulation (FANTOM5, ENCODE and GTEx). The updated version of the GTRD provides an integrated view of transcription regulation through a dedicated web interface with advanced browsing and search capabilities, an integrated genome browser, and table reports by cell types, transcription factors, and genes of interest.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

