The expression of Rpb10, a small subunit common to RNA polymerases, is modulated by the R3H domain-containing Rbs1 protein and the Upf1 helicase
- PMID: 33231687
- PMCID: PMC7708074
- DOI: 10.1093/nar/gkaa1069
The expression of Rpb10, a small subunit common to RNA polymerases, is modulated by the R3H domain-containing Rbs1 protein and the Upf1 helicase
Abstract
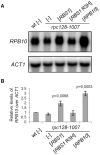
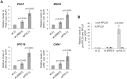
The biogenesis of eukaryotic RNA polymerases is poorly understood. The present study used a combination of genetic and molecular approaches to explore the assembly of RNA polymerase III (Pol III) in yeast. We identified a regulatory link between Rbs1, a Pol III assembly factor, and Rpb10, a small subunit that is common to three RNA polymerases. Overexpression of Rbs1 increased the abundance of both RPB10 mRNA and the Rpb10 protein, which correlated with suppression of Pol III assembly defects. Rbs1 is a poly(A)mRNA-binding protein and mutational analysis identified R3H domain to be required for mRNA interactions and genetic enhancement of Pol III biogenesis. Rbs1 also binds to Upf1 protein, a key component in nonsense-mediated mRNA decay (NMD) and levels of RPB10 mRNA were increased in a upf1Δ strain. Genome-wide RNA binding by Rbs1 was characterized by UV cross-linking based approach. We demonstrated that Rbs1 directly binds to the 3' untranslated regions (3'UTRs) of many mRNAs including transcripts encoding Pol III subunits, Rpb10 and Rpc19. We propose that Rbs1 functions by opposing mRNA degradation, at least in part mediated by NMD pathway. Orthologues of Rbs1 protein are present in other eukaryotes, including humans, suggesting that this is a conserved regulatory mechanism.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures









Similar articles
-
Specific Features of RNA Polymerases I and III: Structure and Assembly.Front Mol Biosci. 2021 May 14;8:680090. doi: 10.3389/fmolb.2021.680090. eCollection 2021. Front Mol Biosci. 2021. PMID: 34055890 Free PMC article. Review.
-
Assembly of RNA polymerase III complex involves a putative co-translational mechanism.Gene. 2022 May 25;824:146394. doi: 10.1016/j.gene.2022.146394. Epub 2022 Mar 9. Gene. 2022. PMID: 35278633 Review.
-
Rbs1 protein, involved in RNA polymerase III complex assembly in the yeast Saccharomyces cerevisiae, induces a Gcn4 response and forms aggregates when overproduced.Gene. 2022 Jan 30;809:146034. doi: 10.1016/j.gene.2021.146034. Epub 2021 Oct 21. Gene. 2022. PMID: 34688816
-
Rbs1, a new protein implicated in RNA polymerase III biogenesis in yeast Saccharomyces cerevisiae.Mol Cell Biol. 2015 Apr;35(7):1169-81. doi: 10.1128/MCB.01230-14. Epub 2015 Jan 20. Mol Cell Biol. 2015. PMID: 25605335 Free PMC article.
-
Reprogramming mRNA Expression in Response to Defect in RNA Polymerase III Assembly in the Yeast Saccharomyces cerevisiae.Int J Mol Sci. 2021 Jul 7;22(14):7298. doi: 10.3390/ijms22147298. Int J Mol Sci. 2021. PMID: 34298922 Free PMC article.
Cited by
-
Internal transcription termination widely regulates differential expression of operon-organized genes including ribosomal protein and RNA polymerase genes in an archaeon.Nucleic Acids Res. 2023 Aug 25;51(15):7851-7867. doi: 10.1093/nar/gkad575. Nucleic Acids Res. 2023. PMID: 37439380 Free PMC article.
-
Never a dull enzyme, RNA polymerase II.Transcription. 2023 Nov;14(1-2):49-67. doi: 10.1080/21541264.2023.2208023. Epub 2023 May 2. Transcription. 2023. PMID: 37132022 Free PMC article. Review.
-
KH domain proteins: Another family of bacterial RNA matchmakers?Mol Microbiol. 2022 Jan;117(1):10-19. doi: 10.1111/mmi.14842. Epub 2021 Nov 19. Mol Microbiol. 2022. PMID: 34748246 Free PMC article. Review.
-
Specific Features of RNA Polymerases I and III: Structure and Assembly.Front Mol Biosci. 2021 May 14;8:680090. doi: 10.3389/fmolb.2021.680090. eCollection 2021. Front Mol Biosci. 2021. PMID: 34055890 Free PMC article. Review.
-
Association between a single nucleotide polymorphism in the R3HCC1 gene and irinotecan toxicity.Cancer Med. 2023 Feb;12(4):4294-4305. doi: 10.1002/cam4.5299. Epub 2022 Oct 29. Cancer Med. 2023. PMID: 36308049 Free PMC article.
References
-
- Werner F., Grohmann D.. Evolution of multisubunit RNA polymerases in the three domains of life. Nat. Rev. Microbiol. 2011; 9:85–98. - PubMed
-
- Abel C.-B., Verónica M.-F., Ana I G.-G., Francisco N.. Subunits common to RNA polymerases. The Yeast Role in Medical Applications. 2017; doi:10.5772/intechopen.70936.
-
- Wild T., Cramer P.. Biogenesis of multisubunit RNA polymerases. Trends Biochem. Sci. 2012; 37:99–105. - PubMed
-
- Khatter H., Vorländer M.K., Müller C.W.. RNA polymerase I and III: similar yet unique. Curr. Opin. Struct. Biol. 2017; 47:88–94. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

