SARS-CoV-2: a new dimension to our understanding of coronaviruses
- PMID: 33231780
- PMCID: PMC7684150
- DOI: 10.1007/s10123-020-00152-y
SARS-CoV-2: a new dimension to our understanding of coronaviruses
Abstract
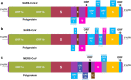
Coronaviruses have marked their significant emergence since the twenty-first century with the outbreaks of three out of the seven existing human coronaviruses, including the severe acute respiratory syndrome coronavirus (SARS-CoV) in 2003, Middle East respiratory syndrome coronavirus (MERS-CoV) in 2012, and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in 2019. These viruses have not only acquired large-scale transmission during their specified outbreak period, but cases of MERS-CoV still remain active, although there is only limited transmission. While, on the other hand, SARS-CoV-2 continues to remain a rising threat to global public health. The recent novel coronavirus, SARS-CoV-2, responsible for the ongoing coronavirus disease 2019 (COVID-19), emerged during December 2019 in Wuhan, China, and has repeatedly raised questions about its characteristic variability. Despite belonging to the same family, SARS-CoV-2 has proven to be quite difficult to control and contain in terms of transmissibility, leading to around 19.8 million reported cases and more than 730,000 deaths of individuals worldwide. Here, we discuss how SARS-CoV-2 differs from its two other related human coronaviruses in terms of genome composition, site of infection, and transmissibility, among several other notable aspects-all indicating to the possibility that it is these variations in addition to other unknowns that are contributing to this virus' differing deadly pattern.
Keywords: ACE2; Case fatality rate; Coronavirus; Genome; MERS; SARS-CoV; SARS-CoV-2; Transmissibility.
Figures
References
-
- Brown AJ, Won JJ, Graham RL, Dinnon KH, III, Sims AC, Feng JY, Cihlar T, Denison MR, Baric RS, Sheahan TP. Broad spectrum antiviral remdesivir inhibits human endemic and zoonotic deltacoronaviruses with a highly divergent RNA dependent RNA polymerase. Antivir Res. 2019;169:104541. doi: 10.1016/j.antiviral.2019.104541. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

