Toward a Molecular Basis of Cellular Nucleoside Transport in Humans
- PMID: 33232132
- PMCID: PMC10032034
- DOI: 10.1021/acs.chemrev.0c00644
Toward a Molecular Basis of Cellular Nucleoside Transport in Humans
Abstract
Nucleosides play central roles in all facets of life, from metabolism to cellular signaling. Because of their physiochemical properties, nucleosides are lipid bilayer impermeable and thus rely on dedicated transport systems to cross biological membranes. In humans, two unrelated protein families mediate nucleoside membrane transport: the concentrative and equilibrative nucleoside transporter families. The objective of this review is to provide a broad outlook on the current status of nucleoside transport research. We will discuss the role played by nucleoside transporters in human health and disease, with emphasis placed on recent structural advancements that have revealed detailed molecular principles of these important cellular transport systems and exploitable pharmacological features.
Conflict of interest statement
Figures


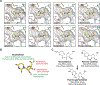

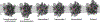
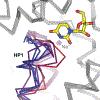
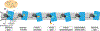

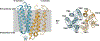
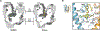
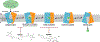

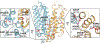

References
-
- Baldwin SA; Mackey JR; Cass CE; Young JD, Nucleoside transporters: Molecular biology and implications for therapeutic development. Mol Med Today 1999, 5, 216–224. - PubMed
-
- King AE; Ackley MA; Cass CE; Young JD; Baldwin SA, Nucleoside transporters: From scavengers to novel therapeutic targets. Trends Pharmacol Sci 2006, 27, 416–425. - PubMed
-
- Parkinson FE; Damaraju VL; Graham K; Yao SYM; Baldwin SA; Cass CE; Young JD, Molecular biology of nucleoside transporters and their distributions and functions in the brain. Curr Top Med Chem 2011, 11, 948–972. - PubMed
-
- Young JD; Yao SYM; Baldwin JM; Cass CE; Baldwin SA, The human concentrative and equilibrative nucleoside transporter families, SLC28 and SLC29. Molecular Aspects of Medicine 2013, 34, 529–547. - PubMed
-
- Young JD; Yao SYM; Sun L; Cass CE; Baldwin SA, Human equilibrative nucleoside transporter (ENT) family of nucleoside and nucleobase transporter proteins. Xenobiotica 2008, 38, 995–1021. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

