Identification of prognostic aging-related genes associated with immunosuppression and inflammation in head and neck squamous cell carcinoma
- PMID: 33232279
- PMCID: PMC7803584
- DOI: 10.18632/aging.104199
Identification of prognostic aging-related genes associated with immunosuppression and inflammation in head and neck squamous cell carcinoma
Abstract
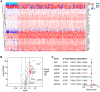
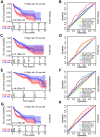
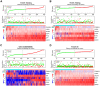
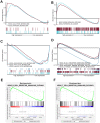
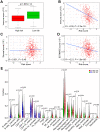
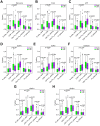
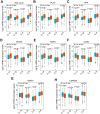
Aging is regarded as a dominant risk factor for cancer. Additionally, inflammation and asthenic immune surveillance with aging may facilitate tumor formation and development. However, few studies have comprehensively analyzed the relationship between aging-related genes (AGs) and the prognosis, inflammation and tumor immunity of head and neck squamous cell carcinoma (HNSCC). Here, we initially screened 41 differentially expressed AGs from The Cancer Genome Atlas (TCGA) database. In the training set, a prognosis risk model with seven AGs (APP, CDKN2A, EGFR, HSPD1, IL2RG, PLAU and VEGFA) was constructed and validated in the TCGA test set and the GEO set (P < 0.05). Using univariate and multivariate Cox regression analyses, we confirmed that risk score was an independent prognostic factor of HNSCC patients. In addition, a high risk score was significantly correlated with immunosuppression, and high expression of PLAU, APP and EGFR was the main factor. Furthermore, we confirmed that a high risk score was significantly associated with levels of proinflammatory factors (IL-1α, IL-1β, IL-6 and IL-8) in HNSCC samples. Thus, this risk model may serve as a prognostic signature and provide clues for individualized immunotherapy for HNSCC patients.
Keywords: aging-related genes (AGs); head and neck squamous cell carcinoma (HNSCC); immunosuppression; inflammation; prognosis.
Conflict of interest statement
Figures


References
-
- Hill SJ, D’Andrea AD. Predictive potential of head and neck squamous cell carcinoma organoids. Cancer Discov. 2019; 9:828–30. 10.1158/2159-8290.CD-19-0527 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

