Discovering novel driver mutations from pan-cancer analysis of mutational and gene expression profiles
- PMID: 33232371
- PMCID: PMC7685479
- DOI: 10.1371/journal.pone.0242780
Discovering novel driver mutations from pan-cancer analysis of mutational and gene expression profiles
Abstract
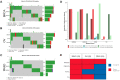
As the genomic profile across cancers varies from person to person, patient prognosis and treatment may differ based on the mutational signature of each tumour. Thus, it is critical to understand genomic drivers of cancer and identify potential mutational commonalities across tumors originating at diverse anatomical sites. Large-scale cancer genomics initiatives, such as TCGA, ICGC and GENIE have enabled the analysis of thousands of tumour genomes. Our goal was to identify new cancer-causing mutations that may be common across tumour sites using mutational and gene expression profiles. Genomic and transcriptomic data from breast, ovarian, and prostate cancers were aggregated and analysed using differential gene expression methods to identify the effect of specific mutations on the expression of multiple genes. Mutated genes associated with the most differentially expressed genes were considered to be novel candidates for driver mutations, and were validated through literature mining, pathway analysis and clinical data investigation. Our driver selection method successfully identified 116 probable novel cancer-causing genes, with 4 discovered in patients having no alterations in any known driver genes: MXRA5, OBSCN, RYR1, and TG. The candidate genes previously not officially classified as cancer-causing showed enrichment in cancer pathways and in cancer diseases. They also matched expectations pertaining to properties of cancer genes, for instance, showing larger gene and protein lengths, and having mutation patterns suggesting oncogenic or tumor suppressor properties. Our approach allows for the identification of novel putative driver genes that are common across cancer sites using an unbiased approach without any a priori knowledge on pathways or gene interactions and is therefore an agnostic approach to the identification of putative common driver genes acting at multiple cancer sites.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

