Defensin-driven viral evolution
- PMID: 33232373
- PMCID: PMC7723274
- DOI: 10.1371/journal.ppat.1009018
Defensin-driven viral evolution
Abstract
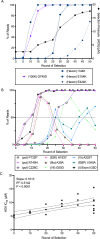
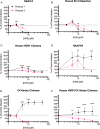
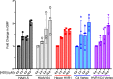
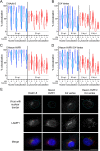
Enteric alpha-defensins are potent effectors of innate immunity that are abundantly expressed in the small intestine. Certain enteric bacteria and viruses are resistant to defensins and even appropriate them to enhance infection despite neutralization of closely related microbes. We therefore hypothesized that defensins impose selective pressure during fecal-oral transmission. Upon passaging a defensin-sensitive serotype of adenovirus in the presence of a human defensin, mutations in the major capsid protein hexon accumulated. In contrast, prior studies identified the vertex proteins as important determinants of defensin antiviral activity. Infection and biochemical assays suggest that a balance between increased cell binding and a downstream block in intracellular trafficking mediated by defensin interactions with all of the major capsid proteins dictates the outcome of infection. These results extensively revise our understanding of the interplay between defensins and non-enveloped viruses. Furthermore, they provide a feasible rationale for defensins shaping viral evolution, resulting in differences in infection phenotypes of closely related viruses.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

