The impact of the Fungus-Host-Microbiota interplay upon Candida albicans infections: current knowledge and new perspectives
- PMID: 33232448
- PMCID: PMC8100220
- DOI: 10.1093/femsre/fuaa060
The impact of the Fungus-Host-Microbiota interplay upon Candida albicans infections: current knowledge and new perspectives
Abstract
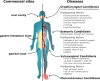
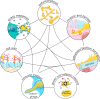
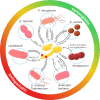
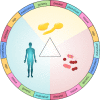
Candida albicans is a major fungal pathogen of humans. It exists as a commensal in the oral cavity, gut or genital tract of most individuals, constrained by the local microbiota, epithelial barriers and immune defences. Their perturbation can lead to fungal outgrowth and the development of mucosal infections such as oropharyngeal or vulvovaginal candidiasis, and patients with compromised immunity are susceptible to life-threatening systemic infections. The importance of the interplay between fungus, host and microbiota in driving the transition from C. albicans commensalism to pathogenicity is widely appreciated. However, the complexity of these interactions, and the significant impact of fungal, host and microbiota variability upon disease severity and outcome, are less well understood. Therefore, we summarise the features of the fungus that promote infection, and how genetic variation between clinical isolates influences pathogenicity. We discuss antifungal immunity, how this differs between mucosae, and how individual variation influences a person's susceptibility to infection. Also, we describe factors that influence the composition of gut, oral and vaginal microbiotas, and how these affect fungal colonisation and antifungal immunity. We argue that a detailed understanding of these variables, which underlie fungal-host-microbiota interactions, will present opportunities for directed antifungal therapies that benefit vulnerable patients.
Keywords: Candida; Candida infections; antifungal immunity; fungal variability; fungus-host-microbiota interactions; microbiota; microbiota variability; mycobiota; patient variability.
© The Author(s) 2020. Published by Oxford University Press on behalf of FEMS.
Figures




References
-
- Acosta-Rodriguez EV, Rivino L, Geginat Jet al. Surface phenotype and antigenic specificity of human interleukin 17-producing T helper memory cells. Nat Immunol. 2007;8:639–46. - PubMed
-
- Ahrné S, Nobaek S, Jeppsson Bet al. The normal Lactobacillus flora of healthy human rectal and oral mucosa. J Appl Microbiol. 1998;85:88–94. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

