BICRA, a SWI/SNF Complex Member, Is Associated with BAF-Disorder Related Phenotypes in Humans and Model Organisms
- PMID: 33232675
- PMCID: PMC7820627
- DOI: 10.1016/j.ajhg.2020.11.003
BICRA, a SWI/SNF Complex Member, Is Associated with BAF-Disorder Related Phenotypes in Humans and Model Organisms
Abstract
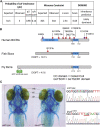
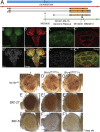
SWI/SNF-related intellectual disability disorders (SSRIDDs) are rare neurodevelopmental disorders characterized by developmental disability, coarse facial features, and fifth digit/nail hypoplasia that are caused by pathogenic variants in genes that encode for members of the SWI/SNF (or BAF) family of chromatin remodeling complexes. We have identified 12 individuals with rare variants (10 loss-of-function, 2 missense) in the BICRA (BRD4 interacting chromatin remodeling complex-associated protein) gene, also known as GLTSCR1, which encodes a subunit of the non-canonical BAF (ncBAF) complex. These individuals exhibited neurodevelopmental phenotypes that include developmental delay, intellectual disability, autism spectrum disorder, and behavioral abnormalities as well as dysmorphic features. Notably, the majority of individuals lack the fifth digit/nail hypoplasia phenotype, a hallmark of most SSRIDDs. To confirm the role of BICRA in the development of these phenotypes, we performed functional characterization of the zebrafish and Drosophila orthologs of BICRA. In zebrafish, a mutation of bicra that mimics one of the loss-of-function variants leads to craniofacial defects possibly akin to the dysmorphic facial features seen in individuals harboring putatively pathogenic BICRA variants. We further show that Bicra physically binds to other non-canonical ncBAF complex members, including the BRD9/7 ortholog, CG7154, and is the defining member of the ncBAF complex in flies. Like other SWI/SNF complex members, loss of Bicra function in flies acts as a dominant enhancer of position effect variegation but in a more context-specific manner. We conclude that haploinsufficiency of BICRA leads to a unique SSRIDD in humans whose phenotypes overlap with those previously reported.
Keywords: BAFopathy; CG11873; Drosophila; GLTSCR1; chromatin; developmental delay; intellectual disability; ncBAF complex; position effect variegation; zebrafish.
Copyright © 2020 American Society of Human Genetics. All rights reserved.
Conflict of interest statement
The Department of Molecular & Human Genetics at Baylor College of Medicine receives revenue from clinical genetic testing conducted at Baylor Genetics Laboratories. C.K. is the Scientific Founder, fiduciary Board of Directors member, Scientific Advisory Board member, shareholder, and consultant of Foghorn Therapeutics, Inc. (Cambridge, MA).
Figures


References
-
- Sexton T., Schober H., Fraser P., Gasser S.M. Gene regulation through nuclear organization. Nat. Struct. Mol. Biol. 2007;14:1049–1055. - PubMed
-
- Ringrose L., Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu. Rev. Genet. 2004;38:413–443. - PubMed
-
- Mohn F., Schübeler D. Genetics and epigenetics: stability and plasticity during cellular differentiation. Trends Genet. 2009;25:129–136. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

