Metagenomic Information Recovery from Human Stool Samples Is Influenced by Sequencing Depth and Profiling Method
- PMID: 33233349
- PMCID: PMC7700633
- DOI: 10.3390/genes11111380
Metagenomic Information Recovery from Human Stool Samples Is Influenced by Sequencing Depth and Profiling Method
Abstract
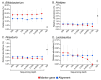
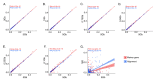
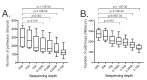
Sequencing of the 16S rRNA gene (16S) has long been a go-to method for microbiome characterization due to its accessibility and lower cost compared to shotgun metagenomic sequencing (SMS). However, 16S sequencing rarely provides species-level resolution and cannot provide direct assessment of other taxa (e.g., viruses and fungi) or functional gene content. Shallow shotgun metagenomic sequencing (SSMS) has emerged as an approach to bridge the gap between 16S sequencing and deep metagenomic sequencing. SSMS is cost-competitive with 16S sequencing, while also providing species-level resolution and functional gene content insights. In the present study, we evaluated the effects of sequencing depth on marker gene-mapping- and alignment-based annotation of bacteria in healthy human stool samples. The number of identified taxa decreased with lower sequencing depths, particularly with the marker gene-mapping-based approach. Other annotations, including viruses and pathways, also showed a depth-dependent effect on feature recovery. These results refine the understanding of the suitability and shortcomings of SSMS, as well as annotation tools for metagenomic analyses in human stool samples. Results may also translate to other sample types and may open the opportunity to explore the effect of sequencing depth and annotation method.
Keywords: alignment; marker gene; microbiome; shallow sequencing; shotgun metagenomic sequencing; virome.
Conflict of interest statement
M.C.R. and J.F.P. declare no conflict of interest. T.M.S.-R., A.G., E.A., A.L.R., Z.H. and T.W. are current employees of Diversigen, Inc., a microbiome services company. E.B.H. is a current employee of Diversigen, Inc., owns OraSure stock/stock options, and has received honoraria for speaking at symposia. W.N. is a former Diversigen, Inc. employee. D.K. serves as Senior Scientific Advisor to Diversigen, Inc.
Figures



References
-
- Johnson J.S., Spakowicz D.J., Hong B.Y., Petersen L.M., Demkowicz P., Chen L., Leopold S.R., Hanson B.M., Agresta H.O., Gerstein M., et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat. Commun. 2019 doi: 10.1038/s41467-019-13036-1. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

