ACE2 Interaction Networks in COVID-19: A Physiological Framework for Prediction of Outcome in Patients with Cardiovascular Risk Factors
- PMID: 33233425
- PMCID: PMC7700637
- DOI: 10.3390/jcm9113743
ACE2 Interaction Networks in COVID-19: A Physiological Framework for Prediction of Outcome in Patients with Cardiovascular Risk Factors
Abstract
Background: Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection (coronavirus disease 2019; COVID-19) is associated with adverse outcomes in patients with cardiovascular disease (CVD). The aim of the study was to characterize the interaction between SARS-CoV-2 and Angiotensin-Converting Enzyme 2 (ACE2) functional networks with a focus on CVD.
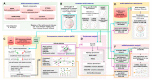
Methods: Using the network medicine approach and publicly available datasets, we investigated ACE2 tissue expression and described ACE2 interaction networks that could be affected by SARS-CoV-2 infection in the heart, lungs and nervous system. We compared them with changes in ACE-2 networks following SARS-CoV-2 infection by analyzing public data of human-induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs). This analysis was performed using the Network by Relative Importance (NERI) algorithm, which integrates protein-protein interaction with co-expression networks. We also performed miRNA-target predictions to identify which miRNAs regulate ACE2-related networks and could play a role in the COVID19 outcome. Finally, we performed enrichment analysis for identifying the main COVID-19 risk groups.
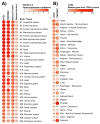
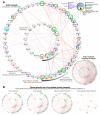
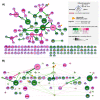
Results: We found similar ACE2 expression confidence levels in respiratory and cardiovascular systems, supporting that heart tissue is a potential target of SARS-CoV-2. Analysis of ACE2 interaction networks in infected hiPSC-CMs identified multiple hub genes with corrupted signaling which can be responsible for cardiovascular symptoms. The most affected genes were EGFR (Epidermal Growth Factor Receptor), FN1 (Fibronectin 1), TP53, HSP90AA1, and APP (Amyloid Beta Precursor Protein), while the most affected interactions were associated with MAST2 and CALM1 (Calmodulin 1). Enrichment analysis revealed multiple diseases associated with the interaction networks of ACE2, especially cancerous diseases, obesity, hypertensive disease, Alzheimer's disease, non-insulin-dependent diabetes mellitus, and congestive heart failure. Among affected ACE2-network components connected with the SARS-Cov-2 interactome, we identified AGT (Angiotensinogen), CAT (Catalase), DPP4 (Dipeptidyl Peptidase 4), CCL2 (C-C Motif Chemokine Ligand 2), TFRC (Transferrin Receptor) and CAV1 (Caveolin-1), associated with cardiovascular risk factors. We described for the first time miRNAs which were common regulators of ACE2 networks and virus-related proteins in all analyzed datasets. The top miRNAs regulating ACE2 networks were miR-27a-3p, miR-26b-5p, miR-10b-5p, miR-302c-5p, hsa-miR-587, hsa-miR-1305, hsa-miR-200b-3p, hsa-miR-124-3p, and hsa-miR-16-5p.
Conclusion: Our study provides a complete mechanistic framework for investigating the ACE2 network which was validated by expression data. This framework predicted risk groups, including the established ones, thus providing reliable novel information regarding the complexity of signaling pathways affected by SARS-CoV-2. It also identified miRNAs that could be used in personalized diagnosis in COVID-19.
Keywords: ACE2; COVID-19; SARS-CoV-2; cardiovascular; gene expression; miR; miRNA; microRNA; therapeutic target.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Thrombosis-related circulating miR-16-5p is associated with disease severity in patients hospitalised for COVID-19.RNA Biol. 2022 Jan;19(1):963-979. doi: 10.1080/15476286.2022.2100629. RNA Biol. 2022. PMID: 35938548 Free PMC article.
-
Alteration of circulating ACE2-network related microRNAs in patients with COVID-19.Sci Rep. 2024 Jun 12;14(1):13573. doi: 10.1038/s41598-024-58037-3. Sci Rep. 2024. PMID: 38866792 Free PMC article.
-
Expression analysis of miRNA hsa-let7b-5p in naso-oropharyngeal swabs of COVID-19 patients supports its role in regulating ACE2 and DPP4 receptors.J Cell Mol Med. 2022 Oct;26(19):4940-4948. doi: 10.1111/jcmm.17492. Epub 2022 Sep 8. J Cell Mol Med. 2022. PMID: 36073344 Free PMC article.
-
Angiotensin-converting enzyme 2 (ACE2), SARS-CoV-2 and the pathophysiology of coronavirus disease 2019 (COVID-19).J Pathol. 2020 Jul;251(3):228-248. doi: 10.1002/path.5471. Epub 2020 Jun 10. J Pathol. 2020. PMID: 32418199 Free PMC article. Review.
-
Angiotensin-Converting Enzyme 2: SARS-CoV-2 Receptor and Regulator of the Renin-Angiotensin System: Celebrating the 20th Anniversary of the Discovery of ACE2.Circ Res. 2020 May 8;126(10):1456-1474. doi: 10.1161/CIRCRESAHA.120.317015. Epub 2020 Apr 8. Circ Res. 2020. PMID: 32264791 Free PMC article. Review.
Cited by
-
SARS-COV-2 as potential microRNA sponge in COVID-19 patients.BMC Med Genomics. 2022 Apr 23;15(Suppl 2):94. doi: 10.1186/s12920-022-01243-7. BMC Med Genomics. 2022. PMID: 35461273 Free PMC article.
-
Host cell entry mediators implicated in the cellular tropism of SARS‑CoV‑2, the pathophysiology of COVID‑19 and the identification of microRNAs that can modulate the expression of these mediators (Review).Int J Mol Med. 2022 Feb;49(2):20. doi: 10.3892/ijmm.2021.5075. Epub 2021 Dec 22. Int J Mol Med. 2022. PMID: 34935057 Free PMC article. Review.
-
Exploring Potential Biomarkers and Molecular Mechanisms of Ischemic Cardiomyopathy and COVID-19 Comorbidity Based on Bioinformatics and Systems Biology.Int J Mol Sci. 2023 Mar 30;24(7):6511. doi: 10.3390/ijms24076511. Int J Mol Sci. 2023. PMID: 37047484 Free PMC article.
-
Heart failure during the COVID-19 pandemic: clinical, diagnostic, management, and organizational dilemmas.ESC Heart Fail. 2022 Dec;9(6):3713-3736. doi: 10.1002/ehf2.14118. Epub 2022 Sep 16. ESC Heart Fail. 2022. PMID: 36111511 Free PMC article.
-
Identification of shared pathogenetic mechanisms between COVID-19 and IC through bioinformatics and system biology.Sci Rep. 2024 Jan 24;14(1):2114. doi: 10.1038/s41598-024-52625-z. Sci Rep. 2024. PMID: 38267482 Free PMC article.
References
-
- Wu C., Chen X., Cai Y., Xia J., Zhou X., Xu S., Huang H., Zhang L., Zhou X., Du C., et al. Risk Factors Associated With Acute Respiratory Distress Syndrome and Death in Patients With Coronavirus Disease 2019 Pneumonia in Wuhan, China. JAMA Intern. Med. 2020;180:934–943. doi: 10.1001/jamainternmed.2020.0994. - DOI - PMC - PubMed
-
- Leung G.M., Hedley A.J., Ho L.-M., Chau P., Wong I.O., Thach T.Q., Ghani A.C., Donnelly C.A., Fraser C., Riley S., et al. The Epidemiology of Severe Acute Respiratory Syndrome in the 2003 Hong Kong Epidemic: An Analysis of All 1755 Patients. Ann. Intern. Med. 2004;141:662–673. doi: 10.7326/0003-4819-141-9-200411020-00006. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

