Classification of Changes in the Fecal Microbiota Associated with Colonic Adenomatous Polyps Using a Long-Read Sequencing Platform
- PMID: 33233735
- PMCID: PMC7699842
- DOI: 10.3390/genes11111374
Classification of Changes in the Fecal Microbiota Associated with Colonic Adenomatous Polyps Using a Long-Read Sequencing Platform
Abstract
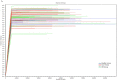
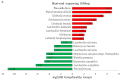
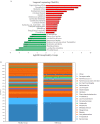
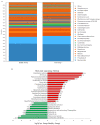
The microbiota is the community of microorganisms that colonizes the oral cavity, respiratory tract, and gut of multicellular organisms. The microbiota exerts manifold physiological and pathological impacts on the organism it inhabits. A growing body of attention is being paid to host-microbiota interplay, which is highly relevant to the development of carcinogenesis. Adenomatous polyps are considered a common hallmark of colorectal cancer, the second leading cause of carcinogenesis-mediated death worldwide. In this study, we examined the relevance between targeted operational taxonomic units and colonic polyps using short- and long-read sequencing platforms. The gut microbiota was assessed in 132 clinical subjects, including 53 healthy participants, 36 patients with occult blood in the gut, and 43 cases with adenomatous polyps. An elevation in the relative abundance of Klebsiella pneumonia, Fusobacterium varium, and Fusobacterium mortiferum was identified in patients with adenomatous polyps compared with the other groups using long-read sequencing workflow. In contrast, the relatively high abundances of Blautia luti, Bacteroides plebeius, and Prevotella copri were characterized in the healthy groups. The diversities in gut microbiota communities were similar in all recruited samples. These results indicated that alterations in gut microbiota were characteristic of participants with adenomatous polyps, which might be relevant to the further development of CRC. These findings provide a potential contribution to the early prediction and interception of CRC occurrence.
Keywords: Oxford nanopore technology; adenomatous polyp; colorectal cancer; gut microbiota.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




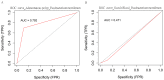
References
-
- Svolos V., Hansen R., Nichols B., Quince C., Ijaz U.Z., Papadopoulou R.T., Edwards C.A., Watson D., Alghamdi A., Brejnrod A., et al. Treatment of Active Crohn’s Disease With an Ordinary Food-based Diet That Replicates Exclusive Enteral Nutrition. Gastroenterology. 2019;156:1354–1367. doi: 10.1053/j.gastro.2018.12.002. - DOI - PubMed
-
- Franzosa E.A., Sirota-Madi A., Avila-Pacheco J., Fornelos N., Haiser H.J., Reinker S., Vatanen T., Hall A.B., Mallick H., McIver L.J., et al. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat. Microbiol. 2019;4:293–305. doi: 10.1038/s41564-018-0306-4. - DOI - PMC - PubMed
-
- Sobhani I., Bergsten E., Couffin S., Amiot A., Nebbad B., Barau C., de’Angelis N., Rabot S., Canoui-Poitrine F., Mestivier D., et al. Colorectal cancer-associated microbiota contributes to oncogenic epigenetic signatures. Proc. Natl. Acad. Sci. USA. 2019;116:24285–24295. doi: 10.1073/pnas.1912129116. - DOI - PMC - PubMed
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical

