Purification and Biochemical Characterization of a New Protease Inhibitor from Conyza dioscoridis with Antimicrobial, Antifungal and Cytotoxic Effects
- PMID: 33233753
- PMCID: PMC7699837
- DOI: 10.3390/molecules25225452
Purification and Biochemical Characterization of a New Protease Inhibitor from Conyza dioscoridis with Antimicrobial, Antifungal and Cytotoxic Effects
Abstract
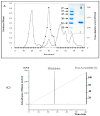
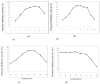
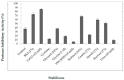
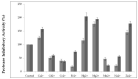
The main objective of the current study was the extraction, purification, and biochemical characterization of a protein protease inhibitor from Conyzadioscoridis. Antimicrobial potential and cytotoxic effects were also examined. The protease inhibitor was extracted in 0.1 M phosphate buffer (pH 6-7). Then, the protease inhibitor, named PDInhibitor, was purified using ammonium sulfate precipitation followed by filtration through a Sephadex G-50 column and had an apparent molecular weight of 25 kDa. The N-terminal sequence of PDInhibitor showed a high level of identity with those of the Kunitz family. PDInhibitor was found to be active at pH values ranging from 5.0 to 11.0, with maximal activity at pH 9.0. It was also fully active at 50 °C and maintained 90% of its stability at over 55 °C. The thermostability of the PDInhibitor was clearly enhanced by CaCl2 and sorbitol, whereas the presence of Ca2+ and Zn2+ ions, Sodium taurodeoxycholate (NaTDC), Sodium dodecyl sulfate (SDS), Dithiothreitol (DTT), and β-ME dramatically improved the inhibitory activity. A remarkable affinity of the protease inhibitor with available important therapeutic proteases (elastase and trypsin) was observed. PDInhibitor also acted as a potent inhibitor of commercial proteases from Aspergillus oryzae and of Proteinase K. The inhibitor displayed potent antimicrobial activity against gram+ and gram- bacteria and against fungal strains. Interestingly, PDInhibitor affected several human cancer cell lines, namely HCT-116, MDA-MB-231, and Lovo. Thus, it can be considered a potentially powerful therapeutic agent.
Keywords: Conyza dioscoridis; characterization antimicrobial effect; protease inhibitor; protein therapeutics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
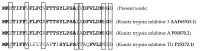




References
-
- Kappelhoff R., Puente X.S., Wilson C.H., Seth A., López-Otín C., Overall C.M. Overview of transcriptomic analysis of all human proteases, non-proteolytic homologs and inhibitors: Organ, tissue and ovarian cancer cell line expression profiling of the human protease degradome by the CLIP-CHIP™ DNA microarray. Biochim. Biophys. Acta (BBA) 2017;1864:2210–2219. doi: 10.1016/j.bbamcr.2017.08.004. - DOI - PubMed
-
- Rawlings N.D., Barrett A.J., Thomas P.D., Huang X., Bateman A., Finn R.D. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2018;46:D624–D632. doi: 10.1093/nar/gkx1134. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

