Common diseases alter the physiological age-related blood microRNA profile
- PMID: 33235214
- PMCID: PMC7686493
- DOI: 10.1038/s41467-020-19665-1
Common diseases alter the physiological age-related blood microRNA profile
Abstract
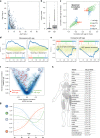
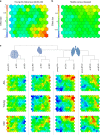
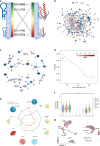
Aging is a key risk factor for chronic diseases of the elderly. MicroRNAs regulate post-transcriptional gene silencing through base-pair binding on their target mRNAs. We identified nonlinear changes in age-related microRNAs by analyzing whole blood from 1334 healthy individuals. We observed a larger influence of the age as compared to the sex and provide evidence for a shift to the 5' mature form of miRNAs in healthy aging. The addition of 3059 diseased patients uncovered pan-disease and disease-specific alterations in aging profiles. Disease biomarker sets for all diseases were different between young and old patients. Computational deconvolution of whole-blood miRNAs into blood cell types suggests that cell intrinsic gene expression changes may impart greater significance than cell abundance changes to the whole blood miRNA profile. Altogether, these data provide a foundation for understanding the relationship between healthy aging and disease, and for the development of age-specific disease biomarkers.
Conflict of interest statement
M.K. is also employed by Hummingbird Diagnostic GmbH. The remaining authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

