Identification of key genes in osteoarthritis using bioinformatics, principal component analysis and meta-analysis
- PMID: 33235627
- PMCID: PMC7678638
- DOI: 10.3892/etm.2020.9450
Identification of key genes in osteoarthritis using bioinformatics, principal component analysis and meta-analysis
Abstract
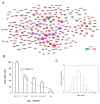
The present study aimed to identify key genes involved in osteoarthritis (OA). Based on a bioinformatics analysis of five gene expression profiling datasets (GSE55457, GSE55235, GSE82107, GSE12021 and GSE1919), differentially expressed genes (DEGs) in OA were identified. Subsequently, a protein-protein interaction (PPI) network was constructed and its topological structure was analyzed. In addition, key genes in OA were identified following a principal component analysis (PCA) based on the DEGs in the PPI network. Finally, the functions and pathways enriched by these key genes were also analyzed. The PPI network consisted of 241 nodes and 576 interactives, including a total of 171 upregulated DEGs [e.g., aspartylglucosaminidase (AGA), CD58 and CD86] and a total of 70 downregulated DEGs (e.g., acetyl-CoA carboxylase β and dihydropyrimidine dehydrogenase). The PPI network complied with an attribute of scale-free small-world network. After PCA, 47 key genes were identified, including β-1,4-galactosyltransferase-1 (B4GALT1), AGA, CD58, CD86, ezrin, and eukaryotic translation initiation factor 4 γ 1 (EIF4G1). Subsequently, the 47 key genes were identified to be enriched in 13 Gene Ontology (GO) terms and 2 Kyoto Encyclopedia of Genes and Genomes pathways, with the GO terms involving B4GALT1 including positive regulation of developmental processes, protein amino acid terminal glycosylation and protein amino acid terminal N-glycosylation. In addition, B4GALT1 and EIF4G1 were confirmed to be downregulated in OA samples compared with healthy controls, but only EIF4G1 was determined to be significantly downregulated in OA samples, as determined via a meta-analysis of the 5 abovementioned datasets. In conclusion, B4GALT1 and EIF4G1 were indicated to have significant roles in OA, and B4GALT1 may be involved in positive regulation of developmental processes, protein amino acid terminal glycosylation and protein amino acid terminal N-glycosylation. The present study may enhance the current understanding of the molecular mechanisms of OA and provide novel therapeutic targets.
Keywords: genomic meta-analysis; network; osteoarthritis; principal component analysis.
Copyright: © Sun et al.
Figures




Similar articles
-
Identification of differential key biomarkers in the synovial tissue between rheumatoid arthritis and osteoarthritis using bioinformatics analysis.Clin Rheumatol. 2021 Dec;40(12):5103-5110. doi: 10.1007/s10067-021-05825-1. Epub 2021 Jul 5. Clin Rheumatol. 2021. PMID: 34224029
-
Gene Expression Microarray Data Identify Hub Genes Involved in Osteoarthritis.Front Genet. 2022 Jun 6;13:870590. doi: 10.3389/fgene.2022.870590. eCollection 2022. Front Genet. 2022. PMID: 35734433 Free PMC article.
-
Identification of a potential gene target for osteoarthritis based on bioinformatics analyses.J Orthop Surg Res. 2020 Jun 22;15(1):228. doi: 10.1186/s13018-020-01756-w. J Orthop Surg Res. 2020. PMID: 32571421 Free PMC article.
-
Identification of Key Genes and Pathways in Osteoarthritis via Bioinformatic Tools: An Updated Analysis.Cartilage. 2021 Dec;13(1_suppl):1457S-1464S. doi: 10.1177/19476035211008975. Epub 2021 Apr 15. Cartilage. 2021. PMID: 33855867 Free PMC article.
-
Identification of Key Genes and Pathways Associated with Sex Differences in Osteoarthritis Based on Bioinformatics Analysis.Biomed Res Int. 2019 Dec 6;2019:3482751. doi: 10.1155/2019/3482751. eCollection 2019. Biomed Res Int. 2019. PMID: 31886203 Free PMC article.
References
-
- Vos T, Allen C, Arora M, Barber RM, Bhutta ZA, Brown A, Carter A, Casey DC, Charlson FJ, Chen AZ, Coggeshall M. Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990-2015: A systematic analysis for the global burden of disease study 2015. Lancet. 2016;388:1545–1602. doi: 10.1016/S0140-6736(16)31678-6. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
