In silico analysis and expression of a new chimeric antigen as a vaccine candidate against cutaneous leishmaniasis
- PMID: 33235698
- PMCID: PMC7671421
- DOI: 10.22038/ijbms.2020.45394.10561
In silico analysis and expression of a new chimeric antigen as a vaccine candidate against cutaneous leishmaniasis
Abstract
Objectives: Since leishmaniasis is one of the health problems in many countries, the development of preventive vaccines against it is a top priority. Peptide vaccines may be a new way to fight the Leishmania infection. In this study, a silicon method was used to predict and analyze B and T cells to produce a vaccine against cutaneous leishmaniasis.
Materials and methods: Immunodominant epitope of Leishmania were selected from four TSA, LPG3, GP63, and Lmsti1 antigens and linked together using a flexible linker (SAPGTP). The antigenic and allergenic features, 2D and 3D structures, and physicochemical features of a chimeric protein were predicted. Finally, through bioinformatics methods, the mRNA structure was predicted and was produced chemically and cloned into the pLEXY-neo2 vector.
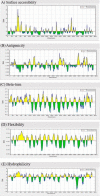
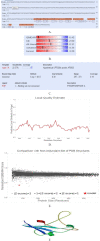
Results: Results indicated, polytope had no allergenic properties, but its antigenicity was estimated to be 0.92%. The amino acids numbers, molecular weight as well as negative and positive charge residuals were estimated 390, ~41KDa, 41, and 30, respectively. The results showed that the designed polytope has 50 post-translationally modified sites. Also, the secondary structure of the protein is composed of 25.38% alpha-helix, 12.31% extended strand, and 62.31% random coil. The results of SDS-PAGE and Western blotting revealed the recombinant protein with ~ 41 kDa. The results of Ramachandran plot showed that 96%, 2.7%, and 1.3% of amino acid residues were located in the preferred, permitted, and outlier areas, respectively.
Conclusion: It is expected that the TLGL polytope will produce a cellular immune response. Therefore, the polytope could be a good candidate for an anti-leishmanial vaccine.
Keywords: Bioinformatics; Leishmania major; Polytope; TLGL; Vaccine.
Figures






References
LinkOut - more resources
Full Text Sources
Research Materials
