Minireview of progress in the structural study of SARS-CoV-2 proteins
- PMID: 33236001
- PMCID: PMC7323663
- DOI: 10.1016/j.crmicr.2020.06.003
Minireview of progress in the structural study of SARS-CoV-2 proteins
Abstract
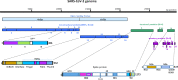
A severe form of pneumonia, named coronavirus disease 2019 (COVID-19) by the World Health Organization, broke out in China and rapidly developed into a global pandemic, with millions of cases and hundreds of thousands of deaths reported globally. The novel coronavirus, which was designated as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), was identified as the etiological agent of COVID-19. On the basis of experience accumulated following previous SARS-CoV and MERS-CoV outbreaks and research, a series of studies have been conducted rapidly, and major progress has been achieved with regard to the understanding of the phylogeny and genomic organization of SARS-CoV-2 in addition its molecular mechanisms of infection and replication. In the present review, we summarized crucial developments in the elucidation of the structure and function of key SARS-CoV-2 proteins, especially the main protease, RNA-dependent RNA polymerase, spike glycoprotein, and nucleocapsid protein. Results of studies on their associated inhibitors and drugs have also been highlighted.
Keywords: 3CLpro, 3C-like protease; 6-HB, six-helix bundle; ACE2, angiotensin-converting enzyme 2; COVID-19; COVID-19, coronavirus disease 2019; CatB/L, cysteine proteases-cathepsin B and L; Drug-screening; E protein, Envelope protein; Genome-encoded proteins; HR1, heptad repeat 1; HR2, heptad repeat 2; M protein, Membrane protein; MERS-CoV, the Middle Eastern respiratory syndrome coronavirus; Mpro, Main protease; N protein, Nucleocapsid protein; NSP, non-structural protein; ORF, Open reading frame; PD, peptidase domain; RBD, receptor-binding domain; RBM, receptor-binding motif; RMP, The remdesivir monophosphate; RdRp, RNA-dependent RNA polymerase; S protein, Spike glycoprotein; SARS-CoV, severe acute respiratory syndrome coronavirus; SARS-CoV-2; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; Structure-based screening; gRNA, genomic RNA; sgRNA, subgenomic RNA.
© 2020 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


Similar articles
-
Single domain antibodies derived from ancient animals as broadly neutralizing agents for SARS-CoV-2 and other coronaviruses.Biomed Eng Adv. 2022 Dec;4:100054. doi: 10.1016/j.bea.2022.100054. Epub 2022 Sep 18. Biomed Eng Adv. 2022. PMID: 36158162 Free PMC article. Review.
-
Current Strategies of Antiviral Drug Discovery for COVID-19.Front Mol Biosci. 2021 May 13;8:671263. doi: 10.3389/fmolb.2021.671263. eCollection 2021. Front Mol Biosci. 2021. PMID: 34055887 Free PMC article. Review.
-
Emergence, evolution, and vaccine production approaches of SARS-CoV-2 virus: Benefits of getting vaccinated and common questions.Saudi J Biol Sci. 2022 Apr;29(4):1981-1997. doi: 10.1016/j.sjbs.2021.12.020. Epub 2021 Dec 13. Saudi J Biol Sci. 2022. PMID: 34924802 Free PMC article. Review.
-
Unlocking COVID therapeutic targets: A structure-based rationale against SARS-CoV-2, SARS-CoV and MERS-CoV Spike.Comput Struct Biotechnol J. 2020 Jul 31;18:2117-2131. doi: 10.1016/j.csbj.2020.07.017. eCollection 2020. Comput Struct Biotechnol J. 2020. PMID: 32913581 Free PMC article.
-
From SARS and MERS CoVs to SARS-CoV-2: Moving toward more biased codon usage in viral structural and nonstructural genes.J Med Virol. 2020 Jun;92(6):660-666. doi: 10.1002/jmv.25754. Epub 2020 Mar 16. J Med Virol. 2020. PMID: 32159237 Free PMC article.
Cited by
-
Pan-coronavirus fusion inhibitors to combat COVID-19 and other emerging coronavirus infectious diseases.J Med Virol. 2023 Jan;95(1):e28143. doi: 10.1002/jmv.28143. Epub 2022 Sep 22. J Med Virol. 2023. PMID: 36098460 Free PMC article. Review.
-
Clinical features and effectiveness of Chinese medicine in patients with COVID-19 from overseas: A retrospective study in Xiamen, China.Front Public Health. 2022 Oct 24;10:1038017. doi: 10.3389/fpubh.2022.1038017. eCollection 2022. Front Public Health. 2022. PMID: 36353282 Free PMC article.
-
Structural Basis of Potential Inhibitors Targeting SARS-CoV-2 Main Protease.Front Chem. 2021 Mar 12;9:622898. doi: 10.3389/fchem.2021.622898. eCollection 2021. Front Chem. 2021. PMID: 33889562 Free PMC article. Review.
-
Investigation of phytochemicals isolated from selected Saudi medicinal plants as natural inhibitors of SARS CoV-2 main protease: In vitro, molecular docking and simulation analysis.Saudi Pharm J. 2024 May;32(5):102023. doi: 10.1016/j.jsps.2024.102023. Epub 2024 Mar 8. Saudi Pharm J. 2024. PMID: 38550333 Free PMC article.
-
[Progress in source tracking of SARS-CoV-2].Nan Fang Yi Ke Da Xue Xue Bao. 2020 Dec 30;40(12):1838-1842. doi: 10.12122/j.issn.1673-4254.2020.12.22. Nan Fang Yi Ke Da Xue Xue Bao. 2020. PMID: 33380405 Free PMC article. Chinese.
References
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

