Computational guided identification of a citrus flavonoid as potential inhibitor of SARS-CoV-2 main protease
- PMID: 33236176
- PMCID: PMC7685905
- DOI: 10.1007/s11030-020-10150-x
Computational guided identification of a citrus flavonoid as potential inhibitor of SARS-CoV-2 main protease
Abstract
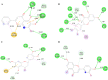
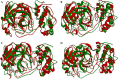
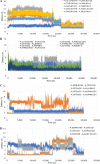
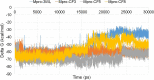
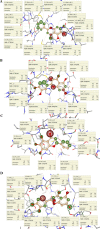
Although vaccine development is being undertaken at a breakneck speed, there is currently no effective antiviral drug for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causing COVID-19. Therefore, the present study aims to explore the possibilities offered by naturally available and abundant flavonoid compounds, as a prospective antiviral drug to combat the virus. A library of 44 citrus flavonoids was screened against the highly conserved Main Protease (Mpro) of SARS-CoV-2 using molecular docking. The compounds which showed better CDocker energy than the co-crystal inhibitor of Mpro were further revalidated by flexible docking within the active site; followed by assessment of drug likeness and toxicity parameters. The non-toxic compounds were further subjected to molecular dynamics simulation and predicted activity (IC50) using 3D-QSAR analysis. Subsequently, hydrogen bonds and dehydration analysis of the best compound were performed to assess the binding affinity to Mpro. It was observed that out of the 44 citrus flavonoids, five compounds showed lower binding energy with Mpro than the co-crystal ligand. Moreover, these compounds also formed H-bonds with two important catalytic residues His41 and Cys145 of the active sites of Mpro. Three compounds which passed the drug likeness filter showed stable conformation during MD simulations. Among these, the lowest predicted IC50 value was observed for Taxifolin. Therefore, this study suggests that Taxifolin, could be a potential inhibitor against SARS-CoV-2 main protease and can be further analysed by in vitro and in vivo experiments for management of the ongoing pandemic.
Keywords: COVID-19; Flavonoids; Main protease; Molecular docking; Molecular dynamic; SARS-CoV-2.
© 2020. Springer Nature Switzerland AG.
Conflict of interest statement
The authors declare that they have are no conflicts of interest.
Figures



References
-
- WHO Coronavirus Disease (COVID-19) Dashboard WHO Coronavirus Disease (COVID-19) Dashboard. https://covid19.who.int/. Accessed 6 Aug 2020
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

