The deubiquitinase USP36 Regulates DNA replication stress and confers therapeutic resistance through PrimPol stabilization
- PMID: 33237263
- PMCID: PMC7736794
- DOI: 10.1093/nar/gkaa1090
The deubiquitinase USP36 Regulates DNA replication stress and confers therapeutic resistance through PrimPol stabilization
Abstract
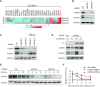
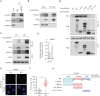
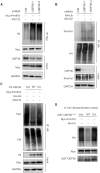
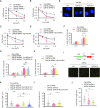
PrimPol has been recently identified as a DNA damage tolerant polymerase that plays an important role in replication stress response. However, the regulatory mechanisms of PrimPol are not well defined. In this study, we identify that the deubiquitinase USP36 interferes with degradation of PrimPol to regulate the replication stress response. Mechanistically, USP36 is deubiquitinated following DNA replication stress, which in turn facilitates its upregulation and interaction with PrimPol. USP36 deubiquitinates K29-linked polyubiquitination of PrimPol and increases its protein stability. Depletion of USP36 results in replication stress-related defects and elevates cell sensitivity to DNA-damage agents, such as cisplatin and olaparib. Moreover, USP36 expression positively correlates with the level of PrimPol protein and poor prognosis in patient samples. These findings indicate that the regulation of PrimPol K29-linked ubiquitination by USP36 plays a critical role in DNA replication stress and chemotherapy response.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




References
-
- Jain R., Aggarwal A.K., Rechkoblit O.. Eukaryotic DNA polymerases. Curr. Opin. Struct. Biol. 2018; 53:77–87. - PubMed
-
- Mouron S., Rodriguez-Acebes S., Martinez-Jimenez M.I., Garcia-Gomez S., Chocron S., Blanco L., Mendez J.. Repriming of DNA synthesis at stalled replication forks by human PrimPol. Nat. Struct. Mol. Biol. 2013; 20:1383–1389. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

