CATH: increased structural coverage of functional space
- PMID: 33237325
- PMCID: PMC7778904
- DOI: 10.1093/nar/gkaa1079
CATH: increased structural coverage of functional space
Abstract
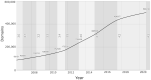
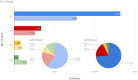
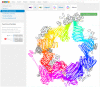
CATH (https://www.cathdb.info) identifies domains in protein structures from wwPDB and classifies these into evolutionary superfamilies, thereby providing structural and functional annotations. There are two levels: CATH-B, a daily snapshot of the latest domain structures and superfamily assignments, and CATH+, with additional derived data, such as predicted sequence domains, and functionally coherent sequence subsets (Functional Families or FunFams). The latest CATH+ release, version 4.3, significantly increases coverage of structural and sequence data, with an addition of 65,351 fully-classified domains structures (+15%), providing 500 238 structural domains, and 151 million predicted sequence domains (+59%) assigned to 5481 superfamilies. The FunFam generation pipeline has been re-engineered to cope with the increased influx of data. Three times more sequences are captured in FunFams, with a concomitant increase in functional purity, information content and structural coverage. FunFam expansion increases the structural annotations provided for experimental GO terms (+59%). We also present CATH-FunVar web-pages displaying variations in protein sequences and their proximity to known or predicted functional sites. We present two case studies (1) putative cancer drivers and (2) SARS-CoV-2 proteins. Finally, we have improved links to and from CATH including SCOP, InterPro, Aquaria and 2DProt.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Orengo C., Michie A., Jones S., Jones D., Swindells M., Thornton J.. CATH – a hierarchic classification of protein domain structures. Structure. 1997; 5:1093–1109. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

