IRBIT activates NBCe1-B by releasing the auto-inhibition module from the transmembrane domain
- PMID: 33237573
- PMCID: PMC7898672
- DOI: 10.1113/JP280578
IRBIT activates NBCe1-B by releasing the auto-inhibition module from the transmembrane domain
Abstract
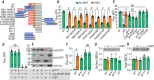
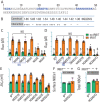
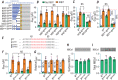
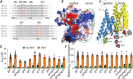
Key points: The electrogenic Na+ /HCO3- cotransporter NBCe1-B is widely expressed in many tissues, including pancreas, submandibular gland, brain, heart, etc. NBCe1-B has very low activity under basal condition due to auto-inhibition, but can be fully activated by protein interaction with the IP3R-binding protein released with inositol 1,4,5-trisphosphate (IRBIT). The structural components of the auto-inhibition domain and the IRBIT-binding domain of NBCe1-B are finely characterized based on systematic mutations in the present study and data from previous studies. Reducing negative charges on the cytosol side of the transmembrane domain greatly decreases the magnitude of the auto-inhibition of NBCe1-B. We propose that the auto-inhibition domain functions as a brake module that inactivates NBCe1-B by binding to, via electrostatic attraction, the transmembrane domain; IRBIT activates NBCe1-B by releasing the brake from the transmembrane domain via competitive binding to the auto-inhibition domain.
Abstract: The electrogenic Na+ /HCO3- cotransporter NBCe1-B is widely expressed in many tissues in the body. NBCe1-B exhibits only basal activity due to the action of the auto-inhibition domain (AID) in its unique amino-terminus. However, NBCe1-B can be activated by interaction with the IP3R-binding protein released with inositol 1,4,5-trisphosphate (IRBIT). Here, we investigate the molecular mechanism underlying the auto-inhibition of NBCe1-B and its activation by IRBIT. The IRBIT-binding domain (IBD) of NBCe1-B spans residues 1-52, essentially consisting of two arms, one negatively charged (residues 1-24) and the other positively charged (residues 40-52). The AID mainly spans residues 40-85, overlapping with the IBD in the positively charged arm. The magnitude of auto-inhibition of NBCe1-B is greatly decreased by manipulating the positively charged residues in the AID or by replacing a set of negatively charged residues with neutral ones in the transmembrane domain. The interaction between IRBIT and NBCe1-B is abolished by mutating a set of negatively charged Asp/Glu residues (to Asn/Gln) plus a set of Ser/Thr residues (to Ala) in the PEST domain of IRBIT. However, this interaction is not affected by replacing the same set of Ser/Thr residues in the PEST domain with Asp. We propose that: (1) the AID, acting as a brake, binds to the transmembrane domain via electrostatic interaction to slow down NBCe1-B; (2) IRBIT activates NBCe1-B by releasing the brake from the transmembrane domain.
Keywords: IRBIT; NBCe1; SLC4A4; auto-inhibition; bicarbonate transporter; electrostatic interaction.
© 2020 The Authors. The Journal of Physiology published by John Wiley & Sons Ltd on behalf of The Physiological Society.
Figures




References
-
- Abuladze N, Lee I, Newman D, Hwang J, Boorer K, Pushkin A & Kurtz I (1998). Molecular cloning, chromosomal localization, tissue distribution, and functional expression of the human pancreatic sodium bicarbonate cotransporter. J Biol Chem 273, 17689–17695. - PubMed
-
- Ando H, Mizutani A, Kiefer H, Tsuzurugi D, Michikawa T & Mikoshiba K (2006). IRBIT suppresses IP3 receptor activity by competing with IP3 for the common binding site on the IP3 receptor. Mol Cell 22, 795–806. - PubMed
-
- Ando H, Mizutani A, Matsu‐ura T & Mikoshiba K (2003). IRBIT, a novel inositol 1,4,5‐trisphosphate (IP3) receptor‐binding protein, is released from the IP3 receptor upon IP3 binding to the receptor. J Biol Chem 278, 10602–10612. - PubMed
-
- Arakawa T, Kobayashi‐Yurugi T, Alguel Y, Iwanari H, Hatae H, Iwata M, Abe Y, Hino T, Ikeda‐Suno C, Kuma H, Kang D, Murata T, Hamakubo T, Cameron AD, Kobayashi T, Hamasaki N & Iwata S (2015). Crystal structure of the anion exchanger domain of human erythrocyte band 3. Science 350, 680–684. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

