Release of Soybean Isoflavones by Using a β-Glucosidase from Alicyclobacillus herbarius
- PMID: 33237595
- PMCID: PMC8048572
- DOI: 10.1002/cbic.202000688
Release of Soybean Isoflavones by Using a β-Glucosidase from Alicyclobacillus herbarius
Abstract
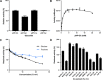
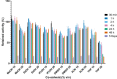
β-Glucosidases are used in the food industry to hydrolyse glycosidic bonds in complex sugars, with enzymes sourced from extremophiles better able to tolerate the process conditions. In this work, a novel β-glycosidase from the acidophilic organism Alicyclobacillus herbarius was cloned and heterologously expressed in Escherichia coli BL21(DE3). AheGH1 was stable over a broad range of pH values (5-11) and temperatures (4-55 °C). The enzyme exhibited excellent tolerance to fructose and good tolerance to glucose, retaining 65 % activity in the presence of 10 % (w/v) glucose. It also tolerated organic solvents, some of which appeared to have a stimulating effect, in particular ethanol with a 1.7-fold increase in activity at 10 % (v/v). The enzyme was then applied for the cleavage of isoflavone from isoflavone glucosides in an ethanolic extract of soy flour, to produce soy isoflavones, which constitute a valuable food supplement, full conversion was achieved within 15 min at 30 °C.
Keywords: biocatalysis; extremophiles; hydrolases; isoflavones; soy.
© 2020 The Authors. ChemBioChem published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Characterization of β-glucosidase from Aspergillus terreus and its application in the hydrolysis of soybean isoflavones.J Zhejiang Univ Sci B. 2016 Jun;17(6):455-64. doi: 10.1631/jzus.B1500317. J Zhejiang Univ Sci B. 2016. PMID: 27256679 Free PMC article.
-
Characterization of a novel isoflavone glycoside-hydrolyzing β-glucosidase from mangrove soil metagenomic library.Biochem Biophys Res Commun. 2021 Sep 10;569:61-65. doi: 10.1016/j.bbrc.2021.06.086. Epub 2021 Jul 3. Biochem Biophys Res Commun. 2021. PMID: 34229124
-
Improve ethanol tolerance of β-glucosidase Bgl1A by semi-rational engineering for the hydrolysis of soybean isoflavone glycosides.J Biotechnol. 2016 Jun 10;227:64-71. doi: 10.1016/j.jbiotec.2016.04.022. Epub 2016 Apr 12. J Biotechnol. 2016. PMID: 27084057
-
A review on applications of β-glucosidase in food, brewery, pharmaceutical and cosmetic industries.Carbohydr Res. 2023 Aug;530:108855. doi: 10.1016/j.carres.2023.108855. Epub 2023 May 25. Carbohydr Res. 2023. PMID: 37263146 Review.
-
Utilization of Isoflavones in Soybeans for Women with Menopausal Syndrome: An Overview.Int J Mol Sci. 2021 Mar 22;22(6):3212. doi: 10.3390/ijms22063212. Int J Mol Sci. 2021. PMID: 33809928 Free PMC article. Review.
Cited by
-
Enzymatic Deglycosylation and Lipophilization of Soy Glycosides into Value-Added Compounds for Food and Cosmetic Applications.ACS Omega. 2025 Mar 20;10(12):12417-12424. doi: 10.1021/acsomega.4c11325. eCollection 2025 Apr 1. ACS Omega. 2025. PMID: 40191329 Free PMC article.
-
Valorization of Apple Pomace: Production of Phloretin Using a Bacterial Cellulose-Immobilized β-Glycosidase.ChemSusChem. 2025 Jul 1;18(13):e202500592. doi: 10.1002/cssc.202500592. Epub 2025 Apr 30. ChemSusChem. 2025. PMID: 40223783 Free PMC article.
-
Production of soy aglycones as an alternative estrogen via innovative synergy of commercial and novel Bacillus coagulans NYO-derived enzymes.Food Chem X. 2025 Jul 4;29:102732. doi: 10.1016/j.fochx.2025.102732. eCollection 2025 Jul. Food Chem X. 2025. PMID: 40686863 Free PMC article.
-
Characterization of β-glucosidase activity of a Lactiplantibacillus plantarum 6-phospho-β-glucosidase.Appl Microbiol Biotechnol. 2025 Apr 8;109(1):86. doi: 10.1007/s00253-025-13472-8. Appl Microbiol Biotechnol. 2025. PMID: 40199767 Free PMC article.
-
Production of Bovine Equol-Enriched Milk: A Review.Animals (Basel). 2021 Mar 8;11(3):735. doi: 10.3390/ani11030735. Animals (Basel). 2021. PMID: 33800327 Free PMC article. Review.
References
-
- Vasella A., Davies G. J., Böhm M., Curr. Opin. Chem. Biol. 2002, 6, 619–629. - PubMed

