Ribosome-Targeting Antibiotics Impair T Cell Effector Function and Ameliorate Autoimmunity by Blocking Mitochondrial Protein Synthesis
- PMID: 33238133
- PMCID: PMC7837214
- DOI: 10.1016/j.immuni.2020.11.001
Ribosome-Targeting Antibiotics Impair T Cell Effector Function and Ameliorate Autoimmunity by Blocking Mitochondrial Protein Synthesis
Abstract
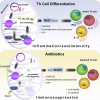
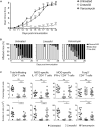
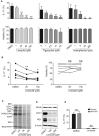
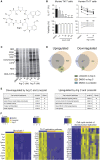
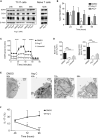
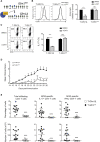
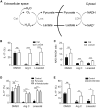
While antibiotics are intended to specifically target bacteria, most are known to affect host cell physiology. In addition, some antibiotic classes are reported as immunosuppressive for reasons that remain unclear. Here, we show that Linezolid, a ribosomal-targeting antibiotic (RAbo), effectively blocked the course of a T cell-mediated autoimmune disease. Linezolid and other RAbos were strong inhibitors of T helper-17 cell effector function in vitro, showing that this effect was independent of their antibiotic activity. Perturbing mitochondrial translation in differentiating T cells, either with RAbos or through the inhibition of mitochondrial elongation factor G1 (mEF-G1) progressively compromised the integrity of the electron transport chain. Ultimately, this led to deficient oxidative phosphorylation, diminishing nicotinamide adenine dinucleotide concentrations and impairing cytokine production in differentiating T cells. In accordance, mice lacking mEF-G1 in T cells were protected from experimental autoimmune encephalomyelitis, demonstrating that this pathway is crucial in maintaining T cell function and pathogenicity.
Keywords: Argyrin; Linezolid; NAD+; T cells; antibiotics; autoimmunity; elongation factor G1; mitochondria; mitochondrial translation; ribosome-targeting.
Copyright © 2020 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures
Comment in
-
Targeting Bacteria within Us to Diminish Infection and Autoimmunity.Immunity. 2021 Jan 12;54(1):1-3. doi: 10.1016/j.immuni.2020.12.006. Immunity. 2021. PMID: 33440134
References
-
- Almeida L., Lochner M., Berod L., Sparwasser T. Metabolic pathways in T cell activation and lineage differentiation. Semin. Immunol. 2016;28:514–524. - PubMed
-
- Appleby R.D., Porteous W.K., Hughes G., James A.M., Shannon D., Wei Y.H., Murphy M.P. Quantitation and origin of the mitochondrial membrane potential in human cells lacking mitochondrial DNA. Eur. J. Biochem. 1999;262:108–116. - PubMed
-
- Armand R., Channon J.Y., Kintner J., White K.A., Miselis K.A., Perez R.P., Lewis L.D. The effects of ethidium bromide induced loss of mitochondrial DNA on mitochondrial phenotype and ultrastructure in a human leukemia T-cell line (MOLT-4 cells) Toxicol. Appl. Pharmacol. 2004;196:68–79. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

