Role of Selective Autophagy in Spermatogenesis and Male Fertility
- PMID: 33238415
- PMCID: PMC7700316
- DOI: 10.3390/cells9112523
Role of Selective Autophagy in Spermatogenesis and Male Fertility
Abstract
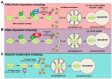
Autophagy is a "self-eating" process that engulfs cellular contents for their subsequent digestion in lysosomes to engage the metabolic need in response to starvation or environmental insults. According to the contents of degradation, autophagy can be divided into bulk autophagy (non-selective autophagy) and selective autophagy. Bulk autophagy degrades non-specific cytoplasmic materials in response to nutrient starvation while selective autophagy targets specific cargoes, such as damaged organelles, protein aggregates, and intracellular pathogens. Selective autophagy has been documented to relate to the reproductive processes, especially for the spermatogenesis, fertilization, and biosynthesis of testosterone. Although selective autophagy is vital in the field of reproduction, its role and the underlying mechanism have remained unclear. In this review, we focus on selective autophagy to discuss the recent advances in our understanding of the mechanism and role of selective autophagy on spermatogenesis and male fertility in mammals. Understanding the role of selective autophagy during spermatogenesis will promote the recognition of genetic regulation in male infertility, and shed light on therapies of infertile patients.
Keywords: autophagy; male fertility; selective autophagy; spermatogenesis.
Conflict of interest statement
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

