GWAS Based on RNA-Seq SNPs and High-Throughput Phenotyping Combined with Climatic Data Highlights the Reservoir of Valuable Genetic Diversity in Regional Tomato Landraces
- PMID: 33238469
- PMCID: PMC7709041
- DOI: 10.3390/genes11111387
GWAS Based on RNA-Seq SNPs and High-Throughput Phenotyping Combined with Climatic Data Highlights the Reservoir of Valuable Genetic Diversity in Regional Tomato Landraces
Abstract
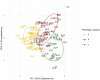
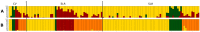
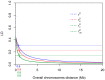
Tomato (Solanum lycopersicum L.) is a widely used model plant species for dissecting out the genomic bases of complex traits to thus provide an optimal platform for modern "-omics" studies and genome-guided breeding. Genome-wide association studies (GWAS) have become a preferred approach for screening large diverse populations and many traits. Here, we present GWAS analysis of a collection of 115 landraces and 11 vintage and modern cultivars. A total of 26 conventional descriptors, 40 traits obtained by digital phenotyping, the fruit content of six carotenoids recorded at the early ripening (breaker) and red-ripe stages and 21 climate-related variables were analyzed in the context of genetic diversity monitored in the 126 accessions. The data obtained from thorough phenotyping and the SNP diversity revealed by sequencing of ripe fruit transcripts of 120 of the tomato accessions were jointly analyzed to determine which genomic regions are implicated in the expressed phenotypic variation. This study reveals that the use of fruit RNA-Seq SNP diversity is effective not only for identification of genomic regions that underlie variation in fruit traits, but also of variation related to additional plant traits and adaptive responses to climate variation. These results allowed validation of our approach because different marker-trait associations mapped on chromosomal regions where other candidate genes for the same traits were previously reported. In addition, previously uncharacterized chromosomal regions were targeted as potentially involved in the expression of variable phenotypes, thus demonstrating that our tomato collection is a precious reservoir of diversity and an excellent tool for gene discovery.
Keywords: RNA-Seq; Solanum lycopersicum L.; digital phenotyping; genome-wide association study (GWAS); genomic diversity; landraces.
Conflict of interest statement
The authors declare that they have no conflicts of interest or competing interests.
Figures
References
-
- Harlan J.R. Our vanishing genetic resources. Science. 1975;188:617–621. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

