Validation of Omega Subunit of RNA Polymerase as a Functional Entity
- PMID: 33238579
- PMCID: PMC7700224
- DOI: 10.3390/biom10111588
Validation of Omega Subunit of RNA Polymerase as a Functional Entity
Abstract
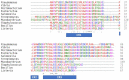
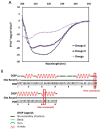
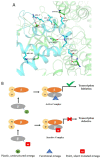
The bacterial RNA polymerase (RNAP) is a multi-subunit protein complex (α2ββ'ω σ) containing the smallest subunit, ω. Although identified early in RNAP research, its function remained ambiguous and shrouded with controversy for a considerable period. It was shown before that the protein has a structural role in maintaining the conformation of the largest subunit, β', and its recruitment in the enzyme assembly. Despite evolutionary conservation of ω and its role in the assembly of RNAP, E. coli mutants lacking rpoZ (codes for ω) are viable due to the association of the global chaperone protein GroEL with RNAP. To get a better insight into the structure and functional role of ω during transcription, several dominant lethal mutants of ω were isolated. The mutants showed higher binding affinity compared to that of native ω to the α2ββ' subassembly. We observed that the interaction between α2ββ' and these lethal mutants is driven by mostly favorable enthalpy and a small but unfavorable negative entropy term. However, during the isolation of these mutants we isolated a silent mutant serendipitously, which showed a lethal phenotype. Silent mutant of a given protein is defined as a protein having the same sequence of amino acids as that of wild type but having mutation in the gene with alteration in base sequence from more frequent code to less frequent one due to codon degeneracy. Eventually, many silent mutants were generated to understand the role of rare codons at various positions in rpoZ. We observed that the dominant lethal mutants of ω having either point mutation or silent in nature are more structured in comparison to the native ω. However, the silent code's position in the reading frame of rpoZ plays a role in the structural alteration of the translated protein. This structural alteration in ω makes it more rigid, which affects the plasticity of the interacting domain formed by ω and α2ββ'. Here, we attempted to describe how the conformational flexibility of the ω helps in maintaining the plasticity of the active site of RNA polymerase. The dominant lethal mutant of ω has a suppressor mapped near the catalytic center of the β' subunit, and it is the same for both types of mutants.
Keywords: RNA polymerase; plasticity; silent mutants; structure; ω-subunit.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Russo F.D., Silhavy T.J. Alpha: The Cinderella subunit of RNA polymerase. J. Biol. Chem. 1992;267:14515–14518. - PubMed
-
- Burgess R.R. Separation and characterization of the subunits of ribonucleic acid polymerase. J. Biol. Chem. 1969;244:6168–6176. - PubMed
-
- Gross C.A., Lonetto M., Losick R. Bacterial sigma factors. In: McKnight S.L., Yamamoto K.R., editors. Transcriptional Regulation. Cold Spring Harbor Laboratory Press; New York, NY, USA: 1992. pp. 129–178.
-
- Weiss S.B., Gladstone L. A mammalian system for the incorporation of cytidine triphosphate into ribonucleic acid. J. Am. Chem. Soc. 1959;81:4118–4119. doi: 10.1021/ja01524a087. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

