The XPB Subunit of the TFIIH Complex Plays a Critical Role in HIV-1 Transcription and XPB Inhibition by Spironolactone Prevents HIV-1 Reactivation from Latency
- PMID: 33239456
- PMCID: PMC7851559
- DOI: 10.1128/JVI.01247-20
The XPB Subunit of the TFIIH Complex Plays a Critical Role in HIV-1 Transcription and XPB Inhibition by Spironolactone Prevents HIV-1 Reactivation from Latency
Abstract
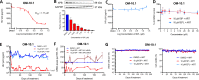
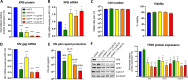
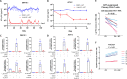
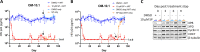
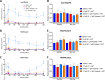
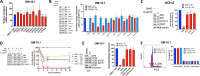
HIV transcription requires assembly of cellular transcription factors at the HIV-1promoter. The TFIIH general transcription factor facilitates transcription initiation by opening the DNA strands around the transcription start site and phosphorylating the C-terminal domain for RNA polymerase II (RNAPII) for activation. Spironolactone (SP), an FDA approved aldosterone antagonist, triggers the proteasomal degradation of the XPB subunit of TFIIH, and concurrently suppresses acute HIV infection in vitro Here we investigated SP as a possible block-and-lock agent for a functional cure aimed at the transcriptional silencing of the viral reservoir. The long-term activity of SP was investigated in primary and cell line models of HIV-1 latency and reactivation. We show that SP rapidly inhibits HIV-1 transcription by reducing RNAPII recruitment to the HIV-1 genome. shRNA knockdown of XPB confirmed XPB degradation as the mechanism of action. Unfortunately, long-term pre-treatment with SP does not result in epigenetic suppression of HIV upon SP treatment interruption, since virus rapidly rebounds when XPB reemerges; however, SP alone without ART maintains the transcriptional suppression. Importantly, SP inhibits HIV reactivation from latency in both cell line models and resting CD4+T cells isolated from aviremic infected individuals upon cell stimulation with latency reversing agents. Furthermore, long-term treatment with concentrations of SP that potently degrade XPB does not lead to global dysregulation of cellular mRNA expression. Overall, these results suggest that XPB plays a key role in HIV transcriptional regulation and XPB degradation by SP strengthens the potential of HIV transcriptional inhibitors in block-and-lock HIV cure approaches.IMPORTANCE Antiretroviral therapy (ART) effectively reduces an individual's HIV loads to below the detection limit, nevertheless rapid viral rebound immediately ensues upon treatment interruption. Furthermore, virally suppressed individuals experience chronic immune activation from ongoing low-level virus expression. Thus, the importance of identifying novel therapeutics to explore in block-and-lock HIV functional cure approaches, aimed at the transcriptional and epigenetic silencing of the viral reservoir to block reactivation from latency. We investigated the potential of repurposing the FDA-approved spironolactone (SP), as one such drug. SP treatment rapidly degrades a host transcription factor subunit, XPB, inhibiting HIV transcription and blocking reactivation from latency. Long-term SP treatment does not affect cellular viability, cell cycle progression or global cellular transcription. SP alone blocks HIV transcription in the absence of ART but does not delay rebound upon drug removal as XPB rapidly reemerges. This study highlights XPB as a novel drug target in block-and-lock therapeutic approaches.
Copyright © 2020 American Society for Microbiology.
Figures


References
-
- Chun TW, Carruth L, Finzi D, Shen X, DiGiuseppe JA, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn TC, Kuo YH, Brookmeyer R, Zeiger MA, Barditch-Crovo P, Siliciano RF. 1997. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 387:183–188. doi: 10.1038/387183a0. - DOI - PubMed
-
- Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, Quinn TC, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho DD, Richman DD, Siliciano RF. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295–1300. doi: 10.1126/science.278.5341.1295. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

