Genomic retargeting of p53 and CTCF is associated with transcriptional changes during oncogenic HRas-induced transformation
- PMID: 33239721
- PMCID: PMC7809021
- DOI: 10.1038/s42003-020-01398-y
Genomic retargeting of p53 and CTCF is associated with transcriptional changes during oncogenic HRas-induced transformation
Abstract
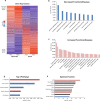
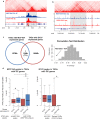
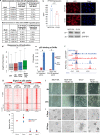
Gene transcription is regulated by distant regulatory elements via combinatorial binding of transcription factors. It is increasingly recognized that alterations in chromatin state and transcription factor binding in these distant regulatory elements may have key roles in cancer development. Here we focused on the first stages of oncogene-induced carcinogenic transformation, and characterized the regulatory network underlying transcriptional changes associated with this process. Using Hi-C data, we observe spatial coupling between differentially expressed genes and their differentially accessible regulatory elements and reveal two candidate transcription factors, p53 and CTCF, as determinants of transcriptional alterations at the early stages of oncogenic HRas-induced transformation in human mammary epithelial cells. Strikingly, the malignant transcriptional reprograming is promoted by redistribution of chromatin binding of these factors without major variation in their expression level. Our results demonstrate that alterations in the regulatory landscape have a major role in driving oncogene-induced transcriptional reprogramming.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

