TRF2-independent chromosome end protection during pluripotency
- PMID: 33239783
- PMCID: PMC7614352
- DOI: 10.1038/s41586-020-2960-y
TRF2-independent chromosome end protection during pluripotency
Abstract
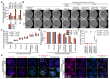
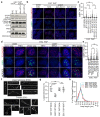
Mammalian telomeres protect chromosome ends from aberrant DNA repair1. TRF2, a component of the telomere-specific shelterin protein complex, facilitates end protection through sequestration of the terminal telomere repeat sequence within a lariat T-loop structure2,3. Deleting TRF2 (also known as TERF2) in somatic cells abolishes T-loop formation, which coincides with telomere deprotection, chromosome end-to-end fusions and inviability3-9. Here we establish that, by contrast, TRF2 is largely dispensable for telomere protection in mouse pluripotent embryonic stem (ES) and epiblast stem cells. ES cell telomeres devoid of TRF2 instead activate an attenuated telomeric DNA damage response that lacks accompanying telomere fusions, and propagate for multiple generations. The induction of telomere dysfunction in ES cells, consistent with somatic deletion of Trf2 (also known as Terf2), occurs only following the removal of the entire shelterin complex. Consistent with TRF2 being largely dispensable for telomere protection specifically during early embryonic development, cells exiting pluripotency rapidly switch to TRF2-dependent end protection. In addition, Trf2-null embryos arrest before implantation, with evidence of strong DNA damage response signalling and apoptosis specifically in the non-pluripotent compartment. Finally, we show that ES cells form T-loops independently of TRF2, which reveals why TRF2 is dispensable for end protection during pluripotency. Collectively, these data establish that telomere protection is solved by distinct mechanisms in pluripotent and somatic tissues.
Conflict of interest statement
Author Information
All data are archived at the Francis Crick Institute or CMRI. The authors declare no competing interests. Requests for materials should be addressed to
Figures
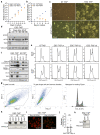




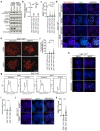


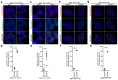
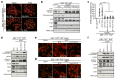

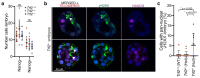
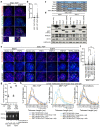
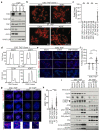

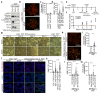
References
-
- Palm W, de Lange T. How shelterin protects mammalian telomeres. Annual review of genetics. 2008;42:301–334. - PubMed
-
- Griffith JDC, C L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Mammalian Telomeres End in a Large Duplex Loop. Cell. 1999;97:503–514. - PubMed
-
- van Steensel B, S A, de Lange T. TRF2 Protects Human Telomeres from End-to-End Fusions. Cell. 1998;92:401–413. - PubMed
-
- Karlseder J, B D, Dai Y, Hardy S, de Lange T. p53- and ATM-Dependent Apoptosis Induced by Telomeres Lacking TRF2. Science. 1999;283:1321–1325. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

