TRF2-mediated telomere protection is dispensable in pluripotent stem cells
- PMID: 33239785
- PMCID: PMC9161009
- DOI: 10.1038/s41586-020-2959-4
TRF2-mediated telomere protection is dispensable in pluripotent stem cells
Abstract
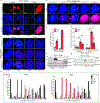
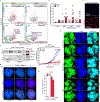
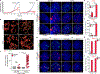
In mammals, telomere protection is mediated by the essential protein TRF2, which binds chromosome ends and ensures genome integrity1,2. TRF2 depletion results in end-to-end chromosome fusions in all cell types that have been tested so far. Here we find that TRF2 is dispensable for the proliferation and survival of mouse embryonic stem (ES) cells. Trf2-/- (also known as Terf2) ES cells do not exhibit telomere fusions and can be expanded indefinitely. In response to the deletion of TRF2, ES cells exhibit a muted DNA damage response that is characterized by the recruitment of γH2AX-but not 53BP1-to telomeres. To define the mechanisms that control this unique DNA damage response in ES cells, we performed a CRISPR-Cas9-knockout screen. We found a strong dependency of TRF2-null ES cells on the telomere-associated protein POT1B and on the chromatin remodelling factor BRD2. Co-depletion of POT1B or BRD2 with TRF2 restores a canonical DNA damage response at telomeres, resulting in frequent telomere fusions. We found that TRF2 depletion in ES cells activates a totipotent-like two-cell-stage transcriptional program that includes high levels of ZSCAN4. We show that the upregulation of ZSCAN4 contributes to telomere protection in the absence of TRF2. Together, our results uncover a unique response to telomere deprotection during early development.
Conflict of interest statement
Figures




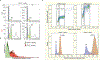



References
-
- Celli GB & de Lange T DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat. Cell Biol. 7, 712–718 (2005). - PubMed
-
- de Lange T Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 19, 2100–2110 (2005). - PubMed
-
- Artandi SE et al. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature 406, 641–645 (2000). - PubMed
-
- Griffith JD et al. Mammalian telomeres end in a large duplex loop. Cell 97, 503–514 (1999). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

