The gut microbiota is associated with immune cell dynamics in humans
- PMID: 33239790
- PMCID: PMC7725892
- DOI: 10.1038/s41586-020-2971-8
The gut microbiota is associated with immune cell dynamics in humans
Abstract
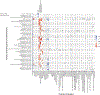
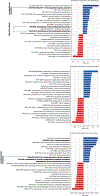
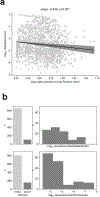
The gut microbiota influences development1-3 and homeostasis4-7 of the mammalian immune system, and is associated with human inflammatory8 and immune diseases9,10 as well as responses to immunotherapy11-14. Nevertheless, our understanding of how gut bacteria modulate the immune system remains limited, particularly in humans, where the difficulty of direct experimentation makes inference challenging. Here we study hundreds of hospitalized-and closely monitored-patients with cancer receiving haematopoietic cell transplantation as they recover from chemotherapy and stem-cell engraftment. This aggressive treatment causes large shifts in both circulatory immune cell and microbiota populations, enabling the relationships between the two to be studied simultaneously. Analysis of observed daily changes in circulating neutrophil, lymphocyte and monocyte counts and more than 10,000 longitudinal microbiota samples revealed consistent associations between gut bacteria and immune cell dynamics. High-resolution clinical metadata and Bayesian inference allowed us to compare the effects of bacterial genera in relation to those of immunomodulatory medications, revealing a considerable influence of the gut microbiota-together and over time-on systemic immune cell dynamics. Our analysis establishes and quantifies the link between the gut microbiota and the human immune system, with implications for microbiota-driven modulation of immunity.
Conflict of interest statement
Competing Interests
MRMvdB and JUP received financial support from Seres Therapeutics. M-AP has received honoraria from AbbVie, Bellicum, Bristol-Myers Squibb, Incyte, Merck, Novartis, Nektar Therapeutics, and Takeda; has received research support for clinical trials from Incyte, Kite (Gilead) and Miltenyi Biotec; and serves on data and safety monitoring boards for Servier and Medigene and scientific advisory boards for MolMed and NexImmune.
Figures


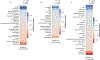







Comment in
-
Tailoring bacterial taxa for immune cell modulation.Hepatobiliary Surg Nutr. 2021 Oct;10(5):686-688. doi: 10.21037/hbsn-21-263. Hepatobiliary Surg Nutr. 2021. PMID: 34760974 Free PMC article. No abstract available.
References
-
- Mazmanian SK, Liu CH, Tzianabos AO & Kasper DL An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122, 107–118 (2005). - PubMed
-
- Gomez de Agüero M et al. The maternal microbiota drives early postnatal innate immune development. Science 351, 1296–1302 (2016). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

