Rxnat: An Open-Source R Package for XNAT-Based Repositories
- PMID: 33240070
- PMCID: PMC7680896
- DOI: 10.3389/fninf.2020.572068
Rxnat: An Open-Source R Package for XNAT-Based Repositories
Abstract
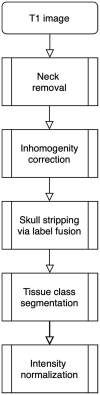
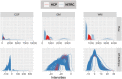
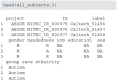
The extensible neuroimaging archive toolkit (XNAT) is a common platform for storing and distributing neuroimaging data and is used by many key repositories of public neuroimaging data. Some examples include the Neuroimaging Informatics Tools and Resources Clearinghouse (NITRC, https://nitrc.org/), the ConnectomeDB for the Human Connectome Project (https://db.humanconnectome.org/), and XNAT Central (https://central.xnat.org/). We introduce Rxnat (https://github.com/adigherman/Rxnat), an open-source R package designed to interact with any XNAT-based repository. The program has similar capabilities with PyXNAT and XNATpy, which were developed for Python users. Rxnat was developed to address the increased popularity of R among neuroimaging researchers. The Rxnat package can query multiple XNAT repositories and download all or a specific subset of images for further processing. This provides a lingua franca for the large community of R analysts to interface with multiple XNAT-based publicly available neuroimaging repositories. The potential of Rxnat is illustrated using an example of neuroimaging data normalization from two neuroimaging repositories, NITRC and HCP.
Keywords: MRI; R; XNAT; connectome; neuroconductor; neuroimaging; nitrc; normalization.
Copyright © 2020 Gherman, Muschelli, Caffo and Crainiceanu.
Figures























Similar articles
-
PyXNAT: XNAT in Python.Front Neuroinform. 2012 May 24;6:12. doi: 10.3389/fninf.2012.00012. eCollection 2012. Front Neuroinform. 2012. PMID: 22654752 Free PMC article.
-
The NITRC image repository.Neuroimage. 2016 Jan 1;124(Pt B):1069-1073. doi: 10.1016/j.neuroimage.2015.05.074. Epub 2015 Jun 2. Neuroimage. 2016. PMID: 26044860 Free PMC article.
-
Data dictionary services in XNAT and the Human Connectome Project.Front Neuroinform. 2014 Jul 3;8:65. doi: 10.3389/fninf.2014.00065. eCollection 2014. Front Neuroinform. 2014. PMID: 25071542 Free PMC article.
-
XNAT Central: Open sourcing imaging research data.Neuroimage. 2016 Jan 1;124(Pt B):1093-1096. doi: 10.1016/j.neuroimage.2015.06.076. Epub 2015 Jul 2. Neuroimage. 2016. PMID: 26143202 Free PMC article.
-
The Extensible Neuroimaging Archive Toolkit: an informatics platform for managing, exploring, and sharing neuroimaging data.Neuroinformatics. 2007 Spring;5(1):11-34. doi: 10.1385/ni:5:1:11. Neuroinformatics. 2007. PMID: 17426351
References
-
- Achterberg H. (2015). XNATpy. Python Module version 0.3.24.
-
- Burger M., Juenemann K., Koenig T. (2018). RUnit: R Unit Test Framework. R package version 0.4.32.
-
- Fisher A. (2020). ggBrain: ggplot Brain Images. R package version 0.1.2.
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

