Advances and opportunities in image analysis of bacterial cells and communities
- PMID: 33242074
- PMCID: PMC8371272
- DOI: 10.1093/femsre/fuaa062
Advances and opportunities in image analysis of bacterial cells and communities
Abstract
The cellular morphology and sub-cellular spatial structure critically influence the function of microbial cells. Similarly, the spatial arrangement of genotypes and phenotypes in microbial communities has important consequences for cooperation, competition, and community functions. Fluorescence microscopy techniques are widely used to measure spatial structure inside living cells and communities, which often results in large numbers of images that are difficult or impossible to analyze manually. The rapidly evolving progress in computational image analysis has recently enabled the quantification of a large number of properties of single cells and communities, based on traditional analysis techniques and convolutional neural networks. Here, we provide a brief introduction to core concepts of automated image processing, recent software tools and how to validate image analysis results. We also discuss recent advances in image analysis of microbial cells and communities, and how these advances open up opportunities for quantitative studies of spatiotemporal processes in microbiology, based on image cytometry and adaptive microscope control.
Keywords: biofilm; data science; machine learning; microbial community; phenotyping; segmentation; single cell.
© The Author(s) 2020. Published by Oxford University Press on behalf of FEMS.
Figures

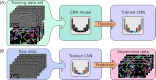

References
-
- Bannon D, Moen E, Borba Eet al. DeepCell 2.0: Automated cloud deployment of deep learning models for large-scale cellular image analysis. bioRxiv. 2018;12:505032.
-
- Becht E, McInnes L, Healy Jet al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol. 2019;37:38–44. - PubMed
-
- Berg HC. How to Track Bacteria. Rev Sci Instrum. 1971;42:868–71. - PubMed

