Excavating the functionally crucial active-site residues of the DXS protein of Bacillus subtilis by exploring its closest homologues
- PMID: 33242110
- PMCID: PMC7691408
- DOI: 10.1186/s43141-020-00087-x
Excavating the functionally crucial active-site residues of the DXS protein of Bacillus subtilis by exploring its closest homologues
Abstract
Background: To achieve a high yield of terpenoid-based therapeutics, 1-deoxy-d-xylulose-5-phosphate (DXP) pathway has been significantly exploited for the production of downstream enzymes. The DXP synthase (DXS) enzyme, the initiator of this pathway, is pivotal for the convergence of carbon flux, and is computationally studied well for the industrially utilized generally regarded as safe (GRAS) bacterium Bacillus subtilis to decode its vital regions for aiding the construction of a functionally improved mutant library.
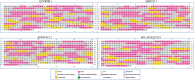
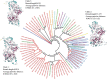
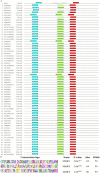
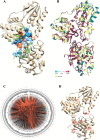
Results: For the 546 sequence dataset of DXS sequences, a representative set of 108 sequences is created, and it shows a significant evolutionary divergence across different species clubbed into 37 clades, whereas three clades are observed for the 76 sequence dataset of Bacillus subtilis. The DXS enzyme, sharing a statistically significant homology to transketolase, is shown to be evolutionarily too distant. By the mutual information-based co-evolutionary network and hotspot analysis, the most crucial loci within the active site are deciphered. The 650-residue representative structure displays a complete conservation of 114 loci, and only two co-evolving residues ASP154 and ILE371 are found to be the conserved ones. Lastly, P318D is predicted to be the top-ranked mutation causing the increase in the thermodynamic stability of 6OUW.
Conclusion: The study excavates the vital functional, phylogenetic, and conserved residues across the active site of the DXS protein, the key rate-limiting controller of the entire pathway. It would aid to computationally understand the evolutionary landscape of this industrially useful enzyme and would allow us to widen its substrate repertoire to increase the enzymatic yield of unnatural molecules for in vivo and in vitro applications.
Keywords: Coevolution; Consurf; DXS; Directed evolution; Motif; Phylogeny.
Conflict of interest statement
None declared.
Figures


Similar articles
-
Computationally Decoding NudF Residues To Enhance the Yield of the DXP Pathway.ACS Omega. 2022 May 27;7(23):19898-19912. doi: 10.1021/acsomega.2c01677. eCollection 2022 Jun 14. ACS Omega. 2022. PMID: 35721994 Free PMC article.
-
Computational analysis of the evolution of the structure and function of 1-deoxy-D-xylulose-5-phosphate synthase, a key regulator of the mevalonate-independent pathway in plants.Gene. 2003 Aug 14;313:127-38. doi: 10.1016/s0378-1119(03)00668-1. Gene. 2003. PMID: 12957384
-
Computational and Synthetic Biology Approaches for the Biosynthesis of Antiviral and Anticancer Terpenoids from Bacillus subtilis.Med Chem. 2022;18(3):307-322. doi: 10.2174/1573406417666210712211557. Med Chem. 2022. PMID: 34254925
-
Phylogenetic analyses and comparative genomics of vitamin B6 (pyridoxine) and pyridoxal phosphate biosynthesis pathways.J Mol Microbiol Biotechnol. 2001 Jan;3(1):1-20. J Mol Microbiol Biotechnol. 2001. PMID: 11200221 Review.
-
DXS as a target for structure-based drug design.Future Med Chem. 2017 Jul;9(11):1277-1294. doi: 10.4155/fmc-2016-0239. Epub 2017 Jun 21. Future Med Chem. 2017. PMID: 28636418 Review.
Cited by
-
Disruption of an Active Site Network Leads to Activation of C2α-Lactylthiamin Diphosphate on the Antibacterial Target 1-Deoxy-d-xylulose-5-phosphate Synthase.Biochemistry. 2024 Mar 5;63(5):671-687. doi: 10.1021/acs.biochem.3c00735. Epub 2024 Feb 23. Biochemistry. 2024. PMID: 38393327 Free PMC article.
-
Potent Inhibition of E. coli DXP Synthase by a gem-Diaryl Bisubstrate Analog.ACS Infect Dis. 2024 Apr 12;10(4):1312-1326. doi: 10.1021/acsinfecdis.3c00734. Epub 2024 Mar 21. ACS Infect Dis. 2024. PMID: 38513073 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
