Dynamic 3D Chromatin Reorganization during Establishment and Maintenance of Pluripotency
- PMID: 33242398
- PMCID: PMC7724465
- DOI: 10.1016/j.stemcr.2020.10.012
Dynamic 3D Chromatin Reorganization during Establishment and Maintenance of Pluripotency
Abstract
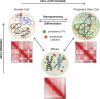
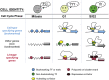
Higher-order chromatin structure is tightly linked to gene expression and therefore cell identity. In recent years, the chromatin landscape of pluripotent stem cells has become better characterized, and unique features at various architectural levels have been revealed. However, the mechanisms that govern establishment and maintenance of these topological characteristics and the temporal and functional relationships with transcriptional or epigenetic features are still areas of intense study. Here, we will discuss progress and limitations of our current understanding regarding how the 3D chromatin topology of pluripotent stem cells is established during somatic cell reprogramming and maintained during cell division. We will also discuss evidence and theories about the driving forces of topological reorganization and the functional links with key features and properties of pluripotent stem cell identity.
Keywords: 3D chromatin organization; ESC; bookmarking; enhancer-promoter interaction; iPSC; mitosis; reprogramming; transcription factors.
Copyright © 2020 The Authors. Published by Elsevier Inc. All rights reserved.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

