Paxillin mediates ATP-induced activation of P2X7 receptor and NLRP3 inflammasome
- PMID: 33243234
- PMCID: PMC7694937
- DOI: 10.1186/s12915-020-00918-w
Paxillin mediates ATP-induced activation of P2X7 receptor and NLRP3 inflammasome
Abstract
Background: Extracellular adenosine triphosphate (ATP), a key danger-associated molecular pattern (DAMP) molecule, is released to the extracellular medium during inflammation by injured parenchymal cells, dying leukocytes, and activated platelets. ATP directly activates the plasma membrane channel P2X7 receptor (P2X7R), leading to an intracellular influx of K+, a key trigger inducing NLRP3 inflammasome activation. However, the mechanism underlying P2X7R-mediated activation of NLRP3 inflammasome is poorly understood, and additional molecular mediators have not been identified. Here, we demonstrate that Paxillin is the molecule connecting the P2X7 receptor and NLRP3 inflammasome through protein interactions.
Results: We show a distinct mechanism by which Paxillin promotes ATP-induced activation of the P2X7 receptor and NLRP3 inflammasome. Extracellular ATP induces Paxillin phosphorylation and then facilitates Paxillin-NLRP3 interaction. Interestingly, Paxillin enhances NLRP3 deubiquitination and activates NLRP3 inflammasome upon ATP treatment and K+ efflux. Moreover, we demonstrated that USP13 is a key enzyme for Paxillin-mediated NLRP3 deubiquitination upon ATP treatment. Notably, extracellular ATP promotes Paxillin and NLRP3 migration from the cytosol to the plasma membrane and facilitates P2X7R-Paxillin interaction and PaxillinNLRP3 association, resulting in the formation of the P2X7R-Paxillin-NLRP3 complex. Functionally, Paxillin is essential for ATP-induced NLRP3 inflammasome activation in mouse BMDMs and BMDCs as well as in human PBMCs and THP-1-differentiated macrophages.
Conclusions: We have identified paxillin as a mediator of NLRP3 inflammasome activation. Paxillin plays key roles in ATP-induced activation of the P2X7 receptor and NLRP3 inflammasome by facilitating the formation of the P2X7R-Paxillin-NLRP3 complex.
Keywords: Adenosine triphosphate, ATP; P2X7 receptor; Paxillin; The NACHT, LRR, and PYD domain-containing protein 3, NLRP3; Ubiquitin-specific peptidase 13, USP13.
Conflict of interest statement
The authors have no conflicts of interest to disclose.
Figures
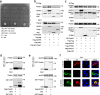







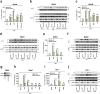

References
Publication types
MeSH terms
Substances
Grants and funding
- 81730061/National Natural Science Foundation of China/International
- 81902053/National Natural Science Foundation of China/International
- 2017ZT07Y580/Guangdong Province Introduction of Innovative R&D Team/International
- 2017ZX10202201/National Health and Family Planning Commission of the People's Republic of China/International
- 2018T110923/China Postdoctoral Science Foundation/International
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

