Direct RNA sequencing reveals m6A modifications on adenovirus RNA are necessary for efficient splicing
- PMID: 33243990
- PMCID: PMC7691994
- DOI: 10.1038/s41467-020-19787-6
Direct RNA sequencing reveals m6A modifications on adenovirus RNA are necessary for efficient splicing
Abstract
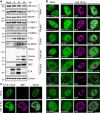
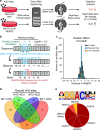
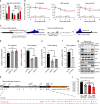
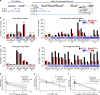
Adenovirus is a nuclear replicating DNA virus reliant on host RNA processing machinery. Processing and metabolism of cellular RNAs can be regulated by METTL3, which catalyzes the addition of N6-methyladenosine (m6A) to mRNAs. While m6A-modified adenoviral RNAs have been previously detected, the location and function of this mark within the infectious cycle is unknown. Since the complex adenovirus transcriptome includes overlapping spliced units that would impede accurate m6A mapping using short-read sequencing, here we profile m6A within the adenovirus transcriptome using a combination of meRIP-seq and direct RNA long-read sequencing to yield both nucleotide and transcript-resolved m6A detection. Although both early and late viral transcripts contain m6A, depletion of m6A writer METTL3 specifically impacts viral late transcripts by reducing their splicing efficiency. These data showcase a new technique for m6A discovery within individual transcripts at nucleotide resolution, and highlight the role of m6A in regulating splicing of a viral pathogen.
Conflict of interest statement
C.E.M. is a cofounder and board member for Biotia and Onegevity Health, as well as an advisor or compensated speaker for Abbvie, Acuamark Diagnostics, ArcBio, BioRad, DNA Genotek, Genialis, Genpro, Karius, Illumina, New England Biolabs, QIAGEN, Whole Biome, and Zymo Research. The remaining authors declare no competing interests.
Figures



References
-
- Arnold J. Berk Adenoviridae. In Fields Virology (eds. David M. Knipe & Peter M. Howley) Vol. 2, 1704–1731 (Wolters Kluwer Health/Lippincott Williams & Wilkins, 2013).
Publication types
MeSH terms
Substances
Grants and funding
- R21 AI147163/AI/NIAID NIH HHS/United States
- R01 AI125416/AI/NIAID NIH HHS/United States
- R01 GM056927/GM/NIGMS NIH HHS/United States
- R01 CA097093/CA/NCI NIH HHS/United States
- R21 AI130618/AI/NIAID NIH HHS/United States
- R01 AI118891/AI/NIAID NIH HHS/United States
- R01 AI152543/AI/NIAID NIH HHS/United States
- F32 AI138432/AI/NIAID NIH HHS/United States
- R01 HD106051/HD/NICHD NIH HHS/United States
- R01 AI145266/AI/NIAID NIH HHS/United States
- R21 AI129851/AI/NIAID NIH HHS/United States
- T32 CA115299/CA/NCI NIH HHS/United States
- R01 AI121321/AI/NIAID NIH HHS/United States
- R01 AI073898/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources

