PcsR2 Is a LuxR-Type Regulator That Is Upregulated on Wheat Roots and Is Unique to Pseudomonas chlororaphis
- PMID: 33244313
- PMCID: PMC7683790
- DOI: 10.3389/fmicb.2020.560124
PcsR2 Is a LuxR-Type Regulator That Is Upregulated on Wheat Roots and Is Unique to Pseudomonas chlororaphis
Abstract
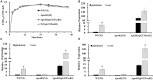
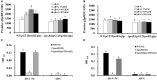
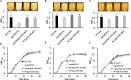
LuxR solos are common in plant-associated bacteria and increasingly recognized for playing important roles in plant-microbe interkingdom signaling. Unlike the LuxR-type transcriptional regulators of prototype LuxR/LuxI quorum sensing systems, luxR solos do not have a LuxI-type autoinducer synthase gene associated with them. LuxR solos in plant-pathogenic bacteria are important for virulence and in plant endosymbionts contribute to symbiosis. In the present study, we characterized an atypical LuxR solo, PcsR2, in the biological control species Pseudomonas chlororaphis 30-84 that is highly conserved among sequenced P. chlororaphis strains. Unlike most LuxR solos in the plant-associated bacteria characterized to date, pcsR2 is not associated with a proline iminopeptidase gene and the protein has an atypical N-terminal binding domain. We created a pcsR2 deletion mutant and used quantitative RT-PCR to show that the expression of pcsR2 and genes in the operon immediately downstream was upregulated ∼10-fold when the wild type strain was grown on wheat roots compared to planktonic culture. PcsR2 was involved in upregulation. Using a GFP transcriptional reporter, we found that expression of pcsR2 responded specifically to root-derived substrates as compared to leaf-derived substrates but not to endogenous AHLs. Compared to the wild type, the mutant was impaired in the ability to utilize root carbon and nitrogen sources in wheat root macerate and to colonize wheat roots. Phenazine production and most biofilm traits previously shown to be correlated with phenazine production also were diminished in the mutant. Gene expression of several of the proteins in the phenazine regulatory network including PhzR, Pip (phenazine inducing protein) and RpeA/RpeB were reduced in the mutant, and overexpression of these genes in trans restored phenazine production in the mutant to wild-type levels, indicating PcsR2 affects the activity of the these regulatory genes upstream of RpeA/RpeB via an undetermined mechanism. Our results indicate PcsR2 upregulates the expression of the adjacent operon in response to unknown wheat root-derived signals and belongs to a novel subfamily of LuxR-type transcriptional regulators found in sequenced P. chlororaphis strains.
Keywords: LuxR solos; Pseudomonas; interkingdom signaling; phenazine; plant-microbe interactions.
Copyright © 2020 Pan, Pierson and Pierson.
Figures

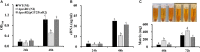
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

