Investigating the human protein-host protein interactome of SARS-CoV-2 infection in the small intestine
- PMID: 33244381
- PMCID: PMC7682973
Investigating the human protein-host protein interactome of SARS-CoV-2 infection in the small intestine
Abstract
Aim: The present study aimed to identify human protein-host protein interactions of SARS-CoV-2 infection in the small intestine to discern the potential mechanisms and gain insights into the associated biomarkers and treatment strategies.
Background: Deciphering the tissue and organ interactions of the SARS-CoV-2 infection can be important to discern the potential underlying mechanisms. In the present study, we investigated the human protein-host protein interactions in the small intestine.
Methods: Public databases and published works were used to collect data related to small intestine tissue and SARS-CoV-2 infection. We constructed a human protein-protein interaction (PPI) network and showed interactions of host proteins in the small intestine. Associated modules, biological processes, functional pathways, regulatory transcription factors, disease ontology categories, and possible drug candidates for therapeutic targets were identified.
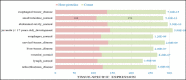
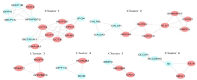
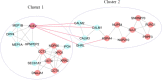
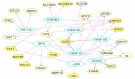
Results: Thirteen primary protein neighbors were found for the SARS-CoV-2 receptor ACE2. ACE2 and its four partners were observed in a highly clustered module; moreover, 8 host proteins belonged to this module. The protein digestion and absorption as a significant pathway was highlighted with enriched genes of ACE2, MEP1A, MEP1B, DPP4, and XPNPEP2. The HNF4A, HNF1A, and HNF1B transcription factors were found to be regulating the expression of ACE2. A significant association with 12 diseases was deciphered and 116 drug-target interactions were identified.
Conclusion: The protein-host protein interactome revealed the important elements and interactions for SARS-CoV-2 infection in the small intestine, which can be useful in clarifying the mechanisms of gastrointestinal symptoms and inflammation. The results suggest that antiviral targeting of these interactions may improve the condition of COVID-19 patients.
Keywords: Drug targets; Interactome; Protein interaction network; Regulatory network; SARS-CoV-2; Small intestine.
©2020 RIGLD, Research Institute for Gastroenterology and Liver Diseases.
Figures



References
-
- Cao W. Clinical features and laboratory inspection of novel coronavirus pneumonia (COVID-19) in Xiangyang, Hubei. medRxiv. 2020
-
- Fang Z, Yi F, Wu K, Lai K, Sun X, Zhong N, et al. Clinical characteristics of 2019 coronavirus pneumonia (COVID-19): an updated systematic review. MedRxiv. 2020
LinkOut - more resources
Full Text Sources
Miscellaneous
