Neisseria gonorrhoeae clustering to reveal major European whole-genome-sequencing-based genogroups in association with antimicrobial resistance
- PMID: 33245688
- PMCID: PMC8208699
- DOI: 10.1099/mgen.0.000481
Neisseria gonorrhoeae clustering to reveal major European whole-genome-sequencing-based genogroups in association with antimicrobial resistance
Abstract
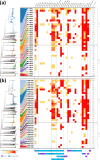
Neisseria gonorrhoeae, the bacterium responsible for the sexually transmitted disease gonorrhoea, has shown an extraordinary ability to develop antimicrobial resistance (AMR) to multiple classes of antimicrobials. With no available vaccine, managing N. gonorrhoeae infections demands effective preventive measures, antibiotic treatment and epidemiological surveillance. The latter two are progressively being supported by the generation of whole-genome sequencing (WGS) data on behalf of national and international surveillance programmes. In this context, this study aims to perform N. gonorrhoeae clustering into genogroups based on WGS data, for enhanced prospective laboratory surveillance. Particularly, it aims to identify the major circulating WGS-genogroups in Europe and to establish a relationship between these and AMR. Ultimately, it enriches public databases by contributing with WGS data from Portuguese isolates spanning 15 years of surveillance. A total of 3791 carefully inspected N. gonorrhoeae genomes from isolates collected across Europe were analysed using a gene-by-gene approach (i.e. using cgMLST). Analysis of cluster composition and stability allowed the classification of isolates into a two-step hierarchical genogroup level determined by two allelic distance thresholds revealing cluster stability. Genogroup clustering in general agreed with available N. gonorrhoeae typing methods [i.e. MLST (multilocus sequence typing), NG-MAST (N. gonorrhoeae multi-antigen sequence typing) and PubMLST core-genome groups], highlighting the predominant genogroups circulating in Europe, and revealed that the vast majority of the genogroups present a dominant AMR profile. Additionally, a non-static gene-by-gene approach combined with a more discriminatory threshold for potential epidemiological linkage enabled us to match data with previous reports on outbreaks or transmission chains. In conclusion, this genogroup assignment allows a comprehensive analysis of N. gonorrhoeae genetic diversity and the identification of the WGS-based genogroups circulating in Europe, while facilitating the assessment (and continuous monitoring) of their frequency, geographical dispersion and potential association with specific AMR signatures. This strategy may benefit public-health actions through the prioritization of genogroups to be controlled, the identification of emerging resistance carriage, and the potential facilitation of data sharing and communication.
Keywords: Neisseria gonorrhoeae; antimicrobial resistance; molecular epidemiology; whole-genome sequencing.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures




References
-
- Newman L, Rowley J, Vander Hoorn S, Wijesooriya NS, Unemo M, et al. Global estimates of the prevalence and incidence of four curable sexually transmitted infections in 2012 based on systematic review and global reporting. PLoS One. 2015;10:e0143304. doi: 10.1371/journal.pone.0143304. - DOI - PMC - PubMed
-
- World Health Organization Report on Global Sexually Transmitted Infection Surveillance, 2018. Geneva: WHO; 2018.
-
- European Centre for Disease Prevention and Control Gonococcal Antimicrobial Susceptibility Surveillance in Europe – Results Summary 2017. Stockholm: ECDC; 2019.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

