Integrative Imaging Reveals SARS-CoV-2-Induced Reshaping of Subcellular Morphologies
- PMID: 33245857
- PMCID: PMC7670925
- DOI: 10.1016/j.chom.2020.11.003
Integrative Imaging Reveals SARS-CoV-2-Induced Reshaping of Subcellular Morphologies
Abstract
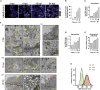
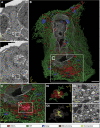
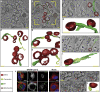
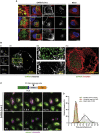
Pathogenesis induced by SARS-CoV-2 is thought to result from both an inflammation-dominated cytokine response and virus-induced cell perturbation causing cell death. Here, we employ an integrative imaging analysis to determine morphological organelle alterations induced in SARS-CoV-2-infected human lung epithelial cells. We report 3D electron microscopy reconstructions of whole cells and subcellular compartments, revealing extensive fragmentation of the Golgi apparatus, alteration of the mitochondrial network and recruitment of peroxisomes to viral replication organelles formed by clusters of double-membrane vesicles (DMVs). These are tethered to the endoplasmic reticulum, providing insights into DMV biogenesis and spatial coordination of SARS-CoV-2 replication. Live cell imaging combined with an infection sensor reveals profound remodeling of cytoskeleton elements. Pharmacological inhibition of their dynamics suppresses SARS-CoV-2 replication. We thus report insights into virus-induced cytopathic effects and provide alongside a comprehensive publicly available repository of 3D datasets of SARS-CoV-2-infected cells for download and smooth online visualization.
Keywords: FIB-SEM; Golgi; coronavirus; cytoskeleton; electron tomography; intermediate filaments; live cell imaging; membrane remodeling; peroxisomes; viral replication organelles.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures


References
-
- Beier T., Pape C., Rahaman N., Prange T., Berg S., Bock D.D., Cardona A., Knott G.W., Plaza S.M., Scheffer L.K. Multicut brings automated neurite segmentation closer to human performance. Nat. Methods. 2017;14:101–102. - PubMed
-
- Chu H., Chan J.F.-W., Yuen T.T.-T., Shuai H., Yuan S., Wang Y., Hu B., Yip C.C.-Y., Tsang J.O.-L., Huang X. Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: an observational study. Lancet Microbe. 2020;1:e14–e23. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

