Both EZH2 and JMJD6 regulate cell cycle genes in breast cancer
- PMID: 33246425
- PMCID: PMC7694428
- DOI: 10.1186/s12885-020-07531-8
Both EZH2 and JMJD6 regulate cell cycle genes in breast cancer
Abstract
Background: Strong evidences support the critical role of Jumonji domain containing 6 (JMJD6) in progression of breast cancer. Here we explore potential partners that coregulate gene expression, to understand additional pathways that are activated by higher amounts of JMJD6.
Methods: We used Gene Set Enrichment Analysis (GSEA) data to identify factors that display gene expression similar to cells treated with JMJD6 siRNA. Using chromatin immunoprecipitations (ChIP) against genomic regions that bind JMJD6 identified by in house and public database Encyclopaedia of DNA Elements (ENCODE), we confirmed JMJD6 occupancy by ChIP PCR. We tested the association of co-regulated genes with patient prognosis using The Cancer Genome Atlas (TCGA) datasets.
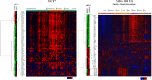
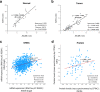
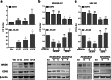
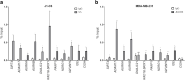
Results: JMJD6 profiles overlapped with those of Enhancer of Zeste homolog 2 (EZH2) and together they appear to co-regulate a unique cassette of genes in both ER+ and ER- cells. 496 genes including aurora kinases, which are currently being tested as novel therapeutic targets in breast cancer were co-regulated in MDA MB 231 cells. JMJD6 and EZH2 neither inter-regulated nor physically interacted with one another. Since both proteins are chromatin modulators, we performed ChIP linked PCR analysis and show that JMJD6 bound in the neighbourhood of co-regulated genes, though EZH2 data did not show any peaks within 100 kb of these sites. Alignment of binding site sequences suggested that atleast two types of binding partners could offer their DNA binding properties to enrich JMJD6 at regulatory sites. In clinical samples, JMJD6 and EZH2 expression significantly correlated in both normal and tumor samples, however the strongest correlation was observed in triple-negative breast cancer (TNBC) subtype. Co-expression of JMJD6 and EZH2 imposed poorer prognosis in breast cancer.
Conclusions: JMJD6 and EZH2 regulate the same crucial cell cycle regulatory and therapeutic targets but their mechanisms appear to be independent of each other. Blocking of a single molecule may not axe cell proliferation completely and blocking both JMJD6 and EZH2 simultaneously may be more effective in breast cancer patients.
Keywords: DREAM; Microarray; Prognosis; TNBC.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Liu Y, Long YH, Wang SQ, Zhang YY, Li YF, Mi JS, Yu CH, Li DY, Zhang JH, Zhang XJ. JMJD6 regulates histone H2A.X phosphorylation and promotes autophagy in triple-negative breast cancer cells via a novel tyrosine kinase activity. Oncogene. 2019;38(7):980–997. doi: 10.1038/s41388-018-0466-y. - DOI - PubMed
-
- Aprelikova O, Chen K, El Touny LH, Brignatz-Guittard C, Han J, Qiu T, Yang HH, Lee MP, Zhu M, Green JE. The epigenetic modifier JMJD6 is amplified in mammary tumors and cooperates with c-Myc to enhance cellular transformation, tumor progression, and metastasis. Clin Epigenetics. 2016;8:38. doi: 10.1186/s13148-016-0205-6. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

