Comprehensive proteomic analysis of exosomes derived from human bone marrow, adipose tissue, and umbilical cord mesenchymal stem cells
- PMID: 33246507
- PMCID: PMC7694919
- DOI: 10.1186/s13287-020-02032-8
Comprehensive proteomic analysis of exosomes derived from human bone marrow, adipose tissue, and umbilical cord mesenchymal stem cells
Abstract
Background: Mesenchymal stem cell (MSC)-derived exosomes have shown comprehensive application prospects over the years. Despite performing similar functions, exosomes from different origins present heterogeneous characteristics and components; however, the relative study remains scarce. Lacking of a valuable reference, researchers select source cells for exosome studies mainly based on accessibility and personal preference.
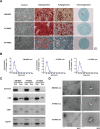
Methods: In this study, exosomes secreted by MSCs derived from different tissues were isolated, by ultracentrifugation, and proteomics analysis was performed. A total of 1014 proteins were detected using a label-free method.
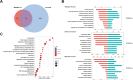
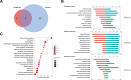
Results: Bioinformatics analysis revealed their shared function in the extracellular matrix receptor. Bone marrow MSC-derived exosomes showed superior regeneration ability, and adipose tissue MSC-derived exosomes played a significant role in immune regulation, whereas umbilical cord MSC-derived exosomes were more prominent in tissue damage repair.
Conclusions: This study systematically and comprehensively analyzes the human MSC-derived exosomes via proteomics, which reveals their potential applications in different fields, so as to provide a reference for researchers to select optimal source cells in future exosome-related studies.
Keywords: Exosomes; Extracellular vesicles; Mesenchymal stem cells; Proteomics; Stem cell-based therapy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

