CRISPR based editing of SIV proviral DNA in ART treated non-human primates
- PMID: 33247091
- PMCID: PMC7695718
- DOI: 10.1038/s41467-020-19821-7
CRISPR based editing of SIV proviral DNA in ART treated non-human primates
Abstract
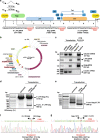
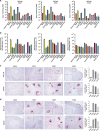
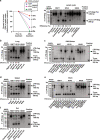
Elimination of HIV DNA from infected individuals remains a challenge in medicine. Here, we demonstrate that intravenous inoculation of SIV-infected macaques, a well-accepted non-human primate model of HIV infection, with adeno-associated virus 9 (AAV9)-CRISPR/Cas9 gene editing construct designed for eliminating proviral SIV DNA, leads to broad distribution of editing molecules and precise cleavage and removal of fragments of the integrated proviral DNA from the genome of infected blood cells and tissues known to be viral reservoirs including lymph nodes, spleen, bone marrow, and brain among others. Accordingly, AAV9-CRISPR treatment results in a reduction in the percent of proviral DNA in blood and tissues. These proof-of-concept observations offer a promising step toward the elimination of HIV reservoirs in the clinic.
Conflict of interest statement
K.K. and R.K. are named inventors on patents that cover the viral gene editing technology that is the subject of this article. In addition to the foregoing interests, K.K. is a co-founder, board member, scientific advisor, and holds equity in Excision Biotherapeutics, a biotech start-up that has licensed the viral gene editing technology from Temple University for commercial development and clinical trials. T.H.B. and J.G. have equity in Excision Biotherapeutics and members of the Scientific Advisory Board. All other authors have no interests to disclose.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

