SARS-CoV-2 genomic and subgenomic RNAs in diagnostic samples are not an indicator of active replication
- PMID: 33247099
- PMCID: PMC7695715
- DOI: 10.1038/s41467-020-19883-7
SARS-CoV-2 genomic and subgenomic RNAs in diagnostic samples are not an indicator of active replication
Abstract
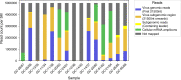
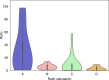
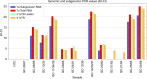
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) was first detected in late December 2019 and has spread worldwide. Coronaviruses are enveloped, positive sense, single-stranded RNA viruses and employ a complicated pattern of virus genome length RNA replication as well as transcription of genome length and leader containing subgenomic RNAs. Although not fully understood, both replication and transcription are thought to take place in so-called double-membrane vesicles in the cytoplasm of infected cells. Here we show detection of SARS-CoV-2 subgenomic RNAs in diagnostic samples up to 17 days after initial detection of infection and provide evidence for their nuclease resistance and protection by cellular membranes suggesting that detection of subgenomic RNAs in such samples may not be a suitable indicator of active coronavirus replication/infection.
Conflict of interest statement
The authors declare no competing or conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

