Genetic requirements and transcriptomics of Helicobacter pylori biofilm formation on abiotic and biotic surfaces
- PMID: 33247117
- PMCID: PMC7695850
- DOI: 10.1038/s41522-020-00167-3
Genetic requirements and transcriptomics of Helicobacter pylori biofilm formation on abiotic and biotic surfaces
Abstract
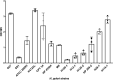
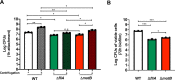
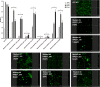
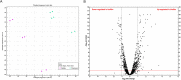
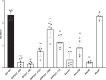
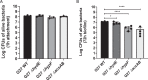
Biofilm growth is a widespread mechanism that protects bacteria against harsh environments, antimicrobials, and immune responses. These types of conditions challenge chronic colonizers such as Helicobacter pylori but it is not fully understood how H. pylori biofilm growth is defined and its impact on H. pylori survival. To provide insights into H. pylori biofilm growth properties, we characterized biofilm formation on abiotic and biotic surfaces, identified genes required for biofilm formation, and defined the biofilm-associated gene expression of the laboratory model H. pylori strain G27. We report that H. pylori G27 forms biofilms with a high biomass and complex flagella-filled 3D structures on both plastic and gastric epithelial cells. Using a screen for biofilm-defective mutants and transcriptomics, we discovered that biofilm cells demonstrated lower transcripts for TCA cycle enzymes but higher ones for flagellar formation, two type four secretion systems, hydrogenase, and acetone metabolism. We confirmed that biofilm formation requires flagella, hydrogenase, and acetone metabolism on both abiotic and biotic surfaces. Altogether, these data suggest that H. pylori is capable of adjusting its phenotype when grown as biofilm, changing its metabolism, and re-shaping flagella, typically locomotion organelles, into adhesive structures.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Helicobacter pylori Biofilm Involves a Multigene Stress-Biased Response, Including a Structural Role for Flagella.mBio. 2018 Oct 30;9(5):e01973-18. doi: 10.1128/mBio.01973-18. mBio. 2018. PMID: 30377283 Free PMC article.
-
Chemorepulsion from the Quorum Signal Autoinducer-2 Promotes Helicobacter pylori Biofilm Dispersal.mBio. 2015 Jul 7;6(4):e00379. doi: 10.1128/mBio.00379-15. mBio. 2015. PMID: 26152582 Free PMC article.
-
The impact of the flagellar protein gene fliK on Helicobacter pylori biofilm formation.mSphere. 2025 Apr 29;10(4):e0001825. doi: 10.1128/msphere.00018-25. Epub 2025 Mar 21. mSphere. 2025. PMID: 40116479 Free PMC article.
-
Helicobacter pylori Biofilm Formation and Its Potential Role in Pathogenesis.Microbiol Mol Biol Rev. 2018 May 9;82(2):e00001-18. doi: 10.1128/MMBR.00001-18. Print 2018 Jun. Microbiol Mol Biol Rev. 2018. PMID: 29743338 Free PMC article. Review.
-
Review article: biofilm formation by Helicobacter pylori as a target for eradication of resistant infection.Aliment Pharmacol Ther. 2012 Aug;36(3):222-30. doi: 10.1111/j.1365-2036.2012.05165.x. Epub 2012 May 31. Aliment Pharmacol Ther. 2012. PMID: 22650647 Review.
Cited by
-
Optimization of Helicobacter pylori Biofilm Formation in In Vitro Conditions Mimicking Stomach.Int J Mol Sci. 2024 Sep 11;25(18):9839. doi: 10.3390/ijms25189839. Int J Mol Sci. 2024. PMID: 39337326 Free PMC article.
-
Temporal dynamics of gene expression during the development of Campylobacter jejuni biofilms.Microb Genom. 2025 May;11(5):001387. doi: 10.1099/mgen.0.001387. Microb Genom. 2025. PMID: 40327030 Free PMC article.
-
Biofilm Formation of Helicobacter pylori in Both Static and Microfluidic Conditions Is Associated With Resistance to Clarithromycin.Front Cell Infect Microbiol. 2022 Mar 25;12:868905. doi: 10.3389/fcimb.2022.868905. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35402304 Free PMC article.
-
Helicobacter pylori in the post-antibiotics era: from virulence factors to new drug targets and therapeutic agents.Arch Microbiol. 2023 Aug 7;205(9):301. doi: 10.1007/s00203-023-03639-0. Arch Microbiol. 2023. PMID: 37550555 Free PMC article. Review.
-
Biofilm of Helicobacter pylori: Life Cycle, Features, and Treatment Options.Antibiotics (Basel). 2023 Jul 31;12(8):1260. doi: 10.3390/antibiotics12081260. Antibiotics (Basel). 2023. PMID: 37627679 Free PMC article. Review.

