Aberrant (pro)renin receptor expression induces genomic instability in pancreatic ductal adenocarcinoma through upregulation of SMARCA5/SNF2H
- PMID: 33247206
- PMCID: PMC7695732
- DOI: 10.1038/s42003-020-01434-x
Aberrant (pro)renin receptor expression induces genomic instability in pancreatic ductal adenocarcinoma through upregulation of SMARCA5/SNF2H
Abstract
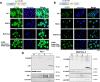
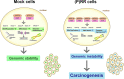
(Pro)renin receptor [(P)RR] has a role in various diseases, such as cardiovascular and renal disorders and cancer. Aberrant (P)RR expression is prevalent in pancreatic ductal adenocarcinoma (PDAC) which is the most common pancreatic cancer. Here we show whether aberrant expression of (P)RR directly leads to genomic instability in human pancreatic ductal epithelial (HPDE) cells. (P)RR-expressing HPDE cells show obvious cellular atypia. Whole genome sequencing reveals that aberrant (P)RR expression induces large numbers of point mutations and structural variations at the genome level. A (P)RR-expressing cell population exhibits tumour-forming ability, showing both atypical nuclei characterised by distinctive nuclear bodies and chromosomal abnormalities. (P)RR overexpression upregulates SWItch/Sucrose Non-Fermentable (SWI/SNF)-related, matrix-associated, actin-dependent regulator of chromatin, subfamily a, member 5 (SMARCA5) through a direct molecular interaction, which results in the failure of several genomic stability pathways. These data reveal that aberrant (P)RR expression contributes to the early carcinogenesis of PDAC.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

