A curated database reveals trends in single-cell transcriptomics
- PMID: 33247933
- PMCID: PMC7698659
- DOI: 10.1093/database/baaa073
A curated database reveals trends in single-cell transcriptomics
Abstract
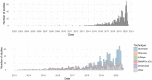
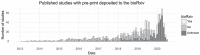
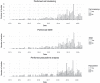
The more than 1000 single-cell transcriptomics studies that have been published to date constitute a valuable and vast resource for biological discovery. While various 'atlas' projects have collated some of the associated datasets, most questions related to specific tissue types, species or other attributes of studies require identifying papers through manual and challenging literature search. To facilitate discovery with published single-cell transcriptomics data, we have assembled a near exhaustive, manually curated database of single-cell transcriptomics studies with key information: descriptions of the type of data and technologies used, along with descriptors of the biological systems studied. Additionally, the database contains summarized information about analysis in the papers, allowing for analysis of trends in the field. As an example, we show that the number of cell types identified in scRNA-seq studies is proportional to the number of cells analysed. Database URL: www.nxn.se/single-cell-studies/gui.
© The Author(s) 2020. Published by Oxford University Press.
Figures


References
-
- Codeluppi S., Borm L.E., Zeisel A., et al. (2018) Spatial organization of the somatosensory cortex revealed by osmFISH. Nat. Methods, 15, 932–935. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

